Automatic Identification and Quantitative Characterization of Primary Dendrite Microstructure Based on Machine Learning
Abstract
:1. Introduction
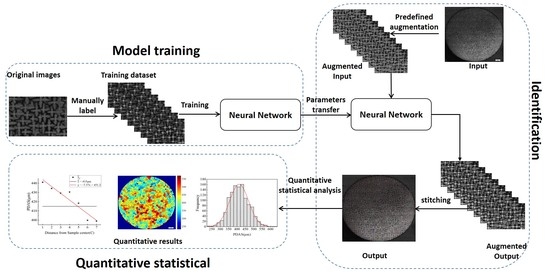
2. Materials and TTA Faster R-CNN Algorithm
2.1. Experimental Data
2.2. TTA Faster R-CNN Algorithm
2.2.1. Faster R-CNN
2.2.2. TTA Faster R-CNN
3. Results
3.1. Automatic Dendrite Core Recognition
3.2. Statistical Results
4. Discussion
5. Conclusions
- (1)
- TTA Faster R-CNN was proposed to automatically select dendrite cores. The method detected 1154 dendritic cores in the single-crystal superalloy metallographic image with a size of 15,360 by 15,360 pixels within 24.5 s, and the recognition accuracy reached 98.4%, which was 15.9% higher than using Faster R-CNN alone.
- (2)
- The local multi-directional algorithm combined with the Voronoi tessellation method is used to calculate the local PDAS of the HT901. There is only a small gap between the average spacing defined by Gaussian distribution of the local PDAS and the average dendritic spacing calculated by the classical counting method, and the difference between the two is only 5 μm.
- (3)
- The technique can quantify the relationship between the local PDAS and the coordination number, and the result suggests that the local PDAS matched by a large coordination number is large.
- (4)
- The mapping of local dendrite spacing to the distance from the center of sample is quantitatively represented by this technique. The result suggests that the closer to the edge, the denser and uneven the dendrite distribution.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HT901 | The name of a self-made second-generation single crystal superalloy. |
| PDAS | Primary Dendrite Arm Spacing. |
| R-CNN | Regions with CNN features-An object detection and semantic segmentation technique. |
| TTA | Test Time Augmentation. |
| TNNs | True Nearest Neighbors. |
| Den Map | Dendritic Mapping-An automatic dendritic mapping algorithm. |
| NCC | Normalized Cross-Correlation. |
| FFT | Fast Fourier Transform. |
| SEM | Scanning Electron Microscopy. |
| RPN | Regional Proposal Network. |
| FCL | Fully Connected Layer. |
| ROI | Region of Interest. |
| TP | True Positive. |
| FN | False Negative. |
| FP | False Positive. |
References
- Strickland, J.; Nenchev, B.; Dong, H. On Directional Dendritic Growth and Primary Spacing—A Review. Crystals 2020, 10, 627. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, L.; Yu, Z.; Zhang, W.; Fu, H. Microstructure development of different orientated nickel-base single crystal superalloy in directional solidification. Mater. Charact. 2010, 61, 7–12. [Google Scholar] [CrossRef]
- Liang, Z.; Chengbo, X.; Guoqing, Z.; Xin, T.; Dingzhong, T. Solidification and Segregation Behavior of Cast Ni-Base Superalloy IN792. Rare Met. Mater. Eng. 2012, 41, 1457–1462. [Google Scholar]
- Brundidge, C.L.; Miller, J.D.; Pollock, T.M. Development of Dendritic Structure in the Liquid-Metal-Cooled, Directional-Solidification Process. Metall. Mater. Trans. A 2011, 42, 2723–2732. [Google Scholar] [CrossRef]
- Mortensen, A.; Cornie, J.A.; Flemings, M.C. Columnar dendritic solidification in a metal- matrix composite. Metall. Trans. A 1988, 19, 709–721. [Google Scholar] [CrossRef]
- Brundidge, C.L.; Van Drasek, D.; Wang, B.; Pollock, T.M. Structure Refinement by a Liquid Metal Cooling Solidification Process for Single-Crystal Nickel-Base Superalloys. Metall. Mater. Trans. A 2012, 43, 965–976. [Google Scholar] [CrossRef]
- Baldan, A.; Magosa, G. Effect of secondary dendrite arm spacing on the section size-dependent creep strength of a nickel-base superalloy. J. Mater. Sci. Lett. 1994, 13, 734–737. [Google Scholar] [CrossRef]
- Krovvidi, S.; Goyal, S.; Bhaduri, A.K. Experimental and Numerical Investigation of High-Temperature Low-Cycle Fatigue and Creep-Fatigue Life of Bellows. J. Mater. Eng. Perform. 2021, 30, 2742–2750. [Google Scholar] [CrossRef]
- Ding, Q.; Bei, H.; Zhao, X.; Gao, Y.; Zhang, Z. Processing, Microstructures and Mechanical Properties of a Ni-Based Single Crystal Superalloy. Crystals 2020, 10, 572. [Google Scholar] [CrossRef]
- Santos, G.A.; Goulart, P.R.; Couto, A.A.; Garcia, A.M.A.U.R.I. Primary Dendrite ARM Spacing Effects upon Mechanical Properties of an AL–3Wt%CU–1Wt%LI Alloy; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Osório, W.R.; Célia Marina AFreire Garcia, A. Dendritic solidification microstructure affecting mechanical and corrosion properties of a Zn4Al alloy. J. Mater. Sci. 2005, 40, 4493–4499. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, L.; Zhang, W.; Yu, Z.; Fu, H. Microstructure and orientation variation during cell/dendrite transition in directional solidification of a single crystal nickel-base superalloy. Mater. Chem. Phys. 2011, 125, 55–58. [Google Scholar] [CrossRef]
- Hui, J.; Tiwari, R.; Wu, X.; Tewari, S.N.; Trivedi, R. Primary dendrite distribution and disorder during directional solidification of Pb-Sb alloys. Metall. Mater. Trans. A 2002, 33, 3499–3510. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Lee, P.D.; Mclean, M. A model of solidification microstructures in nickel-based superalloys: Predicting primary dendrite spacing selection. Acta Mater. 2003, 51, 2971–2987. [Google Scholar] [CrossRef]
- Mccartney, D.G.; Hunt, J.D. Measurements of cell and primary dendrite arm spacings in directionally solidified aluminium alloys. Acta Metall. 1981, 29, 1851–1863. [Google Scholar] [CrossRef]
- Flemings, M.C. Solidification processing. Metall. Trans. 1974, 5, 2121–2134. [Google Scholar] [CrossRef]
- Jacobi, H.; Schwerdtfeger, K. Dendrite morphology of steady state unidirectionally solidified steel. Metall. Trans. A 1976, 7, 811–820. [Google Scholar] [CrossRef]
- Paul, U.; Sahm, P.R.; Goldschmidt, D. Inhomogeneities in single-crystal components. Mater. Sci. Eng. A 1993, 173, 49–54. [Google Scholar] [CrossRef]
- Warnken, N.; Reed, R.C. On the Characterization of Directionally Solidified Dendritic Microstructures. Metall. Mater. Trans. A 2011, 42, 1675–1683. [Google Scholar] [CrossRef]
- Tschopp, M.A.; Miller, J.D.; Oppedal, A.L.; Solanki, K.N. Characterizing the local primary dendrite arm spacing in directionally-solidified dendritic microstructures. Metall. Mater. Trans. A 2013, 45, 426–437. [Google Scholar] [CrossRef] [Green Version]
- Tschopp, M.A.; Groeber, M.A.; Fahringer, R.; Simmons, J.P.; Rosenberger, A.H.; Woodward, C. Automated detection and characterization of microstructural features: Application to eutectic particles in single crystal Ni-based superalloys. Model. Simul. Mater. Sci. Eng. 2010, 18, 025014. [Google Scholar] [CrossRef]
- Wang, Q.; Knight, J.W. Method for Automatic Quantification of Dendrite arm Spacing in Dendritic Microstructures. U.S. Patent No. 9,500,594, 22 November 2016. [Google Scholar]
- Miller, J.; Strangwood, M.; Steinbach, S.; Warnken, N. Skeletonisation to Find the Centre of Dendrites Traced from a 2D Microstructural Image. Solidif. Process. 2017, 2017, 1–4. [Google Scholar]
- Nenchev, B.; Strickland, J.; Tassenberg, K.; Perry, S.; Gill, S.; Dong, H. Automatic Recognition of Dendritic Solidification Structures: Denmap. J. Imaging 2020, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Strickland, J.; Nenchev, B.; Dong, H.B. Applications of pattern recognition for dendritic microstructures. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2020. [Google Scholar]
- Tassenberg, K.; Nenchev, B.; Strickland, J.; Perry, S.; Weston, D. DenMap single crystal solidification structure feature extraction: Automation and application. Mater. Charact. 2020, 171, 110763. [Google Scholar] [CrossRef]
- Yoo, J.C.; Han, T.H. Fast Normalized Cross-Correlation. Circuits Syst. Signal Process. 2009, 28, 819. [Google Scholar] [CrossRef]
- Haque, M.N.; Uddin, M.S. Accelerating Fast Fourier Transformation for Image Processing using Graphics Processing Unit. J. Emerg. Trends Comput. Inf. Sci. 2011, 2, 367–375. [Google Scholar]
- Shanmugam, D.; Blalock, D.; Balakrishnan, G.; Guttag, J. When and Why Test-Time Augmentation Works. arXiv 2020, arXiv:2011.11156. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, X.; Zhang, G.; Ma, S. Deep Learning. Int. J. Semant. Comput. 2016, 10, 417–439. [Google Scholar] [CrossRef] [Green Version]
- Pathirage, C.S.N.; Li, J.; Li, L.; Hao, H.; Liu, W.; Ni, P. Structural damage identification based on autoencoder neural networks and deep learning. Eng. Struct. 2018, 172, 13–28. [Google Scholar] [CrossRef]
- Deng, L.; Yu, D. Deep Learning: Methods and Applications. Found. Trends Signal. Process. 2014, 7, 197–387. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Liu, L.; Zhao, X.; Huang, T.; Yu, Z.; Qu, M.; Fu, H. Effect of cooling rates on dendrite spacings of directionally solidified DZ125 alloy under high thermal gradient. Rare Met. 2009, 28, 633–638. [Google Scholar] [CrossRef]














| Sample Code | Cr | Re | Ta | Al | Co | Mo | W | C | B | Hf | Ni |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HT901 | 5.84 | 2.90 | 5.97 | 5.59 | 8.01 | 2.48 | 4.47 | 0.017 | 0.011 | 0.17 | Bal. |
| Method | TP | FN | FP | Accuracy (%) |
|---|---|---|---|---|
| Faster R-CNN | 964 | 195 | 9 | 82.5 |
| TTA Faster R-CNN | 1147 | 12 | 7 | 98.4 |
| Sample Code | TP | FN | FP | Accuracy (%) |
|---|---|---|---|---|
| HT910 | 1360 | 14 | 27 | 97.1 |
| HT912 | 1237 | 15 | 34 | 96.2 |
| HT913 | 1133 | 17 | 21 | 96.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, W.; Li, D.; Wang, H.; Zhao, L.; Shen, X.; Sun, D.; Chen, J.; Xiao, C. Automatic Identification and Quantitative Characterization of Primary Dendrite Microstructure Based on Machine Learning. Crystals 2021, 11, 1060. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst11091060
Wan W, Li D, Wang H, Zhao L, Shen X, Sun D, Chen J, Xiao C. Automatic Identification and Quantitative Characterization of Primary Dendrite Microstructure Based on Machine Learning. Crystals. 2021; 11(9):1060. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst11091060
Chicago/Turabian StyleWan, Weihao, Dongling Li, Haizhou Wang, Lei Zhao, Xuejing Shen, Dandan Sun, Jingyang Chen, and Chengbo Xiao. 2021. "Automatic Identification and Quantitative Characterization of Primary Dendrite Microstructure Based on Machine Learning" Crystals 11, no. 9: 1060. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst11091060