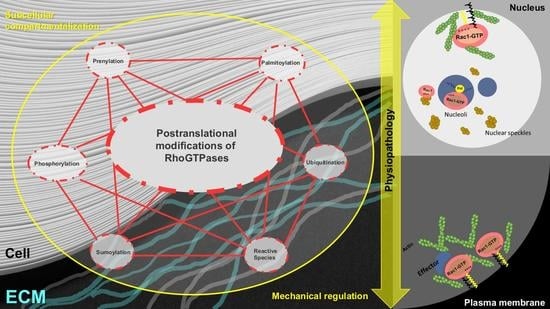
Post-Translational Modification and Subcellular Compartmentalization: Emerging Concepts on the Regulation and Physiopathological Relevance of RhoGTPases
Abstract
:1. Introduction
2. General Aspects of Rho GTPase Modulation
3. PTM of RhoGTPases
3.1. Modification of Rho GTPases by Oxidizing Species
3.2. Phosphorylation of Rho GTPases
3.3. Ubiquitylation and SUMOylation
3.4. Lipid Modifications and Interactions as Modulators of Rho GTPase Subcellular Compartmentalization and Function
3.4.1. Prenylation
3.4.2. Palmitoylation
3.5. Non-Covalent Lipid Interactions
4. Rho GTPases at the Nucleus: Trafficking and Functional Impact
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PTMs | Post-translational modifications |
| GDP/GTP | Guanosine diphosphate and guanosine triphosphate |
| TC | Tumor Cell |
| TME | Tumor microenvironment |
| ECM | Extracellular Matrix |
| CAFs | Cancer Associated Fibroblasts |
| NE | Nuclear Envelope |
| GEFs | Guanine nucleotide Exchange Factors |
| GAPs | GTPase-Activating Proteins |
| GDI | Guanine nucleotide Dissociation Inhibitors |
| EGF | Epithelial Growth Factor |
| SUMO | Small Ubiquitin-like MOdifiers |
| FTase | Farnesyltransferase |
| GGTase I | Geranylgeranyltransferase I |
| PM | Plasma Membrane |
| ZDHHC | Zinc finger DHHC |
| PAT | Palmitoyl-S-Transferase |
| ER | Endoplasmic Reticulum |
| RER | Rugous Endoplasmic Reticulum |
| DRMs | Detergent-Resistant-Membranes |
| CEMMs | Cholesterol Enriched plasma Membrane Microdomains |
| RE | Recycling Endosomes |
| ONM | Outer nuclear membrane |
| INM | Inner-nuclear-membrane |
| PNS | Perinuclear Space |
| NLS | Nuclear Localization Signal |
| NSLS | Nuclear Speckle Localization Signal |
| NES | Nuclear Export Signal |
| LINC | Complex Linker of Nucleoskeleton and Cytoskeleton complex |
References
- Klein, C.A. Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer 2020, 20, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T.; Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009, 10, 243–254. [Google Scholar] [CrossRef]
- Hurst, V.; Shimada, K.; Gasser, S.M. Nuclear actin and actin-binding proteins in DNA Repair. Trends Cell Biol. 2019, 29, 462–476. [Google Scholar] [CrossRef] [Green Version]
- Hall, A. Rho GTPases and the actin cytoskeleton. Science 1998, 279, 509–514. [Google Scholar] [CrossRef] [Green Version]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef]
- Hodge, R.G.; Ridley, A.J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef]
- Popoff, M.R. Bacterial factors exploit eukaryotic Rho GTPase signaling cascades to promote invasion and proliferation within their host. Small GTPases 2014, 5, e983863. [Google Scholar] [CrossRef] [Green Version]
- Kutys, M.L.; Yamada, K.M. Rho GEFs and GAPs: Emerging Integrators of Extracellular Matrix Signaling. Small GTPases 2015, 6, 16–19. [Google Scholar] [CrossRef] [Green Version]
- García-Mata, R.; Wennerberg, K.; Arthur, W.T.; Noren, N.K.; Ellerbroek, S.M.; Burridge, K. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol 2006, 406, 425–437. [Google Scholar] [CrossRef]
- Feig, L.A. Tools of the trade: Use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1999, 1, E25–E27. [Google Scholar] [CrossRef]
- Aspenström, P. Activated Rho GTPases in cancer—the beginning of a new paradigm. Int. J. Mol. Sci. 2018, 19, 3949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, P.M.; Rademacher, J.; Bagshaw, R.D.; Wortmann, C.; Barth, C.; van Unen, J.; Alp, K.M.; Giudice, G.; Eccles, R.L.; Heinrich, L.E.; et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol. 2020, 22, 498–511. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Symons, M.; Campbell, S.L.; Der, C.J. Molecular basis for Rho GTPase signaling specificity. Breast Cancer Res. Treat. 2004, 84, 61–71. [Google Scholar] [CrossRef]
- Garcia-Mata, R.; Boulter, E.; Burridge, K. The “invisible hand”: Regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 2011, 12, 493–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomanbhoy, T.K.; Cerione, R.A. Characterization of the Interaction between RhoGDI and Cdc42Hs Using Fluorescence Spectroscopy. J. Biol. Chem. 1996, 271, 10004–10009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golding, A.E.; Visco, I.; Bieling, P.; Bement, W.M. Extraction of active RhoGTPases by RhoGDI regulates spatiotemporal patterning of RhoGTPases. eLife 2019, 8, e50471. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Mokhtar, A.M.B.; Ahmed, S.B.M.; Darling, N.J.; Harris, M.; Mott, H.R.; Owen, D. A complete survey of RhoGDI targets reveals novel interactions with atypical small GTPases. Biochemistry 2021, 60, 1533–1551. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Bi, F.; Zhou, X.; Zheng, Y. Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol. 2012, 22, 365–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croft, D.R.; Olson, M.F. Transcriptional regulation of Rho GTPase signaling. Transcription 2011, 2, 211–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemichez, E.; Mettouchi, A.; Popoff, M.R. Posttranslational modifications of Rho GTPases mediated by bacterial toxins and cellular systems. In Rho GTPases; Fort, P., Blangy, A., Eds.; World Scientific: Singapore, 2018; pp. 97–118. [Google Scholar]
- Olson, M.F. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases 2018, 9, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS sources in physiological and pathological conditions. Oxidative Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative stress in cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef] [Green Version]
- Forman, H.J.; Fukuto, J.M.; Torres, M. Redox signaling: Thiol chemistry defines which Reactive Oxygen and Nitrogen Species can act as second messengers. Am. J. Physiol. Cell Physiol. 2004, 287, C246–C256. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Zhou, B.; Cox, A.D.; Campbell, S.L. Rho GTPases, oxidation, and cell redox control. Small GTPases 2014, 5, e28579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, J. Redox control of GTPases: From molecular mechanisms to functional significance in health and disease. Antioxid. Redox Signal. 2011, 14, 689–724. [Google Scholar] [CrossRef]
- Heo, J.; Campbell, S.L. Mechanism of redox-mediated guanine nucleotide exchange on redox-active Rho GTPases. J. Biol. Chem. 2005, 280, 31003–31010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghajanian, A.; Wittchen, E.S.; Campbell, S.L.; Burridge, K. Direct activation of RhoA by reactive oxygen species requires a redox-sensitive motif. PLoS ONE 2009, 4, e8045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ushio-Fukai, M. Compartmentalization of redox signaling through NADPH oxidase–derived ROS. Antioxid. Redox Signal. 2009, 11, 1289–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hordijk, P.L. Regulation of NADPH oxidases. Circ. Res. 2006, 98, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Osborn-Heaford, H.L.; Ryan, A.J.; Murthy, S.; Racila, A.-M.; He, C.; Sieren, J.C.; Spitz, D.R.; Carter, A.B. Mitochondrial Rac1 GTPase import and electron transfer from cytochrome c are required for pulmonary fibrosis. J. Biol. Chem. 2012, 287, 3301–3312. [Google Scholar] [CrossRef] [Green Version]
- Nimnual, A.S.; Taylor, L.J.; Bar-Sagi, D. Redox-dependent downregulation of Rho by Rac. Nat. Cell Biol. 2003, 5, 236–241. [Google Scholar] [CrossRef]
- Lang, P.; Gesbert, F.; Delespine-Carmagnat, M.; Stancou, R.; Pouchelet, M.; Bertoglio, J. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 1996, 15, 510–519. [Google Scholar] [CrossRef]
- Smolenski, A.; Lohmann, S.M.; Bertoglio, J.; Chardin, P.; Sauzeau, V.; le Jeune, H.; Cario-Toumaniantz, C.; Pacaud, P.; Loirand, G. Cyclic GMP-dependent protein kinase signaling pathway inhibits RhoA-induced Ca2+ sensitization of contraction in vascular smooth muscle. J. Biol. Chem. 2000, 275, 21722–21729. [Google Scholar] [CrossRef] [Green Version]
- Ellerbroek, S.M.; Wennerberg, K.; Burridge, K. Serine phosphorylation negatively regulates RhoA in vivo. J. Biol. Chem. 2003, 278, 19023–19031. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.; Lemmon, C.; Lietha, D.; Eck, M.; Romer, L. Tyrosine phosphorylation of Rac1: A role in regulation of cell spreading. PLoS ONE 2011, 6, e28587. [Google Scholar] [CrossRef] [Green Version]
- Tu, S.; Wu, W.J.; Wang, J.; Cerione, R.A. Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J. Biol. Chem. 2003, 278, 49293–49300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Ip, J.P.K.; Ye, T.; Ng, Y.-P.; Yung, W.-H.; Wu, Z.; Fang, W.; Fu, A.K.Y.; Ip, N.Y. Cdk5-dependent Mst3 phosphorylation and activity regulate neuronal migration through RhoA inhibition. J. Neurosci. 2014, 34, 7425–7436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, J.; Proff, J.; Hävemeier, A.; Ladwein, M.; Rottner, K.; Barlag, B.; Pich, A.; Tatge, H.; Just, I.; Gerhard, R. Serine-71 phosphorylation of Rac1 modulates downstream signaling. PLoS ONE 2012, 7, e44358. [Google Scholar] [CrossRef] [Green Version]
- Kwon, T.; Kwon, D.Y.; Chun, J.; Kim, J.H.; Kang, S.S. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J. Biol. Chem. 2000, 275, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Abdrabou, A.; Wang, Z. Post-translational modification and subcellular distribution of Rac1: An update. Cells 2018, 7, 263. [Google Scholar] [CrossRef] [Green Version]
- Abdrabou, A.; Brandwein, D.; Liu, C.; Wang, Z. Rac1 S71 mediates the interaction between Rac1 and 14-3-3 proteins. Cells 2019, 8, 1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoentaube, J.; Olling, A.; Tatge, H.; Just, I.; Gerhard, R. Serine-71 phosphorylation of Rac1/Cdc42 diminishes the pathogenic effect of Clostridium difficile toxin A. Cell. Microbiol. 2009, 11, 1816–1826. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation of Rac1 T108 by extracellular signal-regulated kinase in response to epidermal growth factor: A novel mechanism to regulate Rac1 function. Mol. Cell. Biol. 2013, 33, 4538–4551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harreman, M.T.; Kline, T.M.; Milford, H.G.; Harben, M.B.; Hodel, A.E.; Corbett, A.H. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J. Biol. Chem. 2004, 279, 20613–20621. [Google Scholar] [CrossRef] [Green Version]
- Nardozzi, J.D.; Lott, K.; Cingolani, G. Phosphorylation meets nuclear import: A review. Cell Commun. Signal. 2010, 8, 32. [Google Scholar] [CrossRef] [Green Version]
- de la Vega, M.; Burrows, J.F.; Johnston, J.A. Ubiquitination: Added complexity in Ras and Rho family GTPase function. Small GTPases 2011, 2, 192–201. [Google Scholar] [CrossRef] [Green Version]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef] [Green Version]
- Torrino, S.; Visvikis, O.; Doye, A.; Boyer, L.; Stefani, C.; Munro, P.; Bertoglio, J.; Gacon, G.; Mettouchi, A.; Lemichez, E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev. Cell 2011, 21, 959–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majolée, J.; Podieh, F.; Hordijk, P.L.; Kovačević, I. The interplay of Rac1 activity, ubiquitination and GDI binding and its consequences for endothelial cell spreading. PLoS ONE 2021, 16, e0254386. [Google Scholar] [CrossRef]
- Kogler, M.; Tortola, L.; Negri, G.L.; Leopoldi, A.; El-Naggar, A.M.; Mereiter, S.; Gomez-Diaz, C.; Nitsch, R.; Tortora, D.; Kavirayani, A.M.; et al. HACE1 prevents lung carcinogenesis via inhibition of Rac-family GTPases. Cancer Res. 2020, 80, 3009–3022. [Google Scholar] [CrossRef]
- Belaid, A.; Cerezo, M.; Chargui, A.; Corcelle–Termeau, E.; Pedeutour, F.; Giuliano, S.; Ilie, M.; Rubera, I.; Tauc, M.; Barale, S.; et al. Autophagy plays a critical role in the degradation of active RhoA, the control of cell cytokinesis, and genomic stability. Cancer Res. 2013, 73, 4311–4322. [Google Scholar] [CrossRef] [Green Version]
- Choi, B.H.; Chen, C.; Philips, M.; Dai, W. Ras GTPases are modified by SUMOylation. Oncotarget 2018, 9, 4440–4450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, B.H.; Philips, M.R.; Chen, Y.; Lu, L.; Dai, W. K-Ras Lys-42 is crucial for its signaling, cell migration, and invasion. J. Biol. Chem. 2018, 293, 17574–17581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo-Lluva, S.; Tatham, M.H.; Jones, R.C.; Jaffray, E.G.; Edmondson, R.D.; Hay, R.T.; Malliri, A. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat. Cell Biol. 2010, 12, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Lorente, M.; García-Casas, A.; Salvador, N.; Martínez-López, A.; Gabicagogeascoa, E.; Velasco, G.; López-Palomar, L.; Castillo-Lluva, S. Inhibiting SUMO1-mediated SUMOylation induces autophagy-mediated cancer cell death and reduces tumour cell invasion via Rac1. J. Cell Sci. 2019, 132, jcs234120. [Google Scholar] [CrossRef] [Green Version]
- Lao, M.; Zhan, Z.; Li, N.; Xu, S.; Shi, M.; Zou, Y.; Huang, M.; Zeng, S.; Liang, L.; Xu, H. Role of Small Ubiquitin-like MOdifier Proteins-1 (SUMO-1) in regulating migration and invasion of fibroblast-like synoviocytes from patients with rheumatoid arthritis. Exp. Cell Res. 2019, 375, 52–61. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Yu, T.-H.; Wen, W.-L.; Ling, P.; Hsu, K.-S. Conditional deletion of CC2D1A reduces hippocampal synaptic plasticity and impairs cognitive function through Rac1 hyperactivation. J. Neurosci. 2019, 39, 4959–4975. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Khademi, F.; Somesh, B.P.; Rivero, F. Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene 2002, 298, 147–157. [Google Scholar] [CrossRef]
- Zhang, F.L.; Casey, P.J. Protein prenylation: Molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996, 65, 241–269. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M.H.; Brunsveld, L.; Hrycyna, C.A.; Michaelis, S.; Tamanoi, F.; van Voorhis, W.C.; Waldmann, H. Therapeutic intervention based on protein prenylation and associated modifications. Nat. Chem. Biol. 2006, 2, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Casey, P.J. Protein prenylation: Unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 2016, 17, 110–122. [Google Scholar] [CrossRef]
- Furfine, E.S.; Leban, J.J.; Landavazo, A.; Moomaw, J.F.; Casey, P.J. Protein farnesyltransferase: Kinetics of farnesyl pyrophosphate binding and product release. Biochemistry 1995, 34, 6857–6862. [Google Scholar] [CrossRef]
- Taylor, J.S. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 2003, 22, 5963–5974. [Google Scholar] [CrossRef] [Green Version]
- Hicks, K.A.; Hartman, H.L.; Fierke, C.A. Upstream polybasic region in peptides enhances dual specificity for prenylation by both farnesyltransferase and geranyl-geranyltransferase type I. Biochemistry 2005, 44, 15325–15333. [Google Scholar] [CrossRef]
- Winter-Vann, A.M.; Casey, P.J. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 2005, 5, 405–412. [Google Scholar] [CrossRef]
- Young, S.G.; Yang, S.H.; Davies, B.S.J.; Jung, H.-J.; Fong, L.G. Targeting protein prenylation in progeria. Sci. Transl. Med. 2013, 5, 171ps3. [Google Scholar] [CrossRef] [Green Version]
- Storck, E.M.; Morales-Sanfrutos, J.; Serwa, R.A.; Panyain, N.; Lanyon-Hogg, T.; Tolmachova, T.; Ventimiglia, L.N.; Martin-Serrano, J.; Seabra, M.C.; Wojciak-Stothard, B.; et al. Dual chemical probes enable quantitative system-wide analysis of protein prenylation and prenylation dynamics. Nat. Chem. 2019, 11, 552–561. [Google Scholar] [CrossRef]
- Michaelson, D.; Philips, M. The use of GFP to localize Rho GTPases in living cells. Methods Enzymol. 2006, 406, 296–315. [Google Scholar] [CrossRef]
- Roberts, P.J.; Mitin, N.; Keller, P.J.; Chenette, E.J.; Madigan, J.P.; Currin, R.O.; Cox, A.D.; Wilson, O.; Kirschmeier, P.; Der, C.J. Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 2008, 283, 25150–25163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wennerberg, K. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, A.D.; Der, C.J. Ras family signaling: Therapeutic targeting. Cancer Biol. Ther. 2002, 1, 599–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wherlock, M. Farnesyltransferase inhibitors disrupt EGF receptor traffic through modulation of the RhoB GTPase. J. Cell Sci. 2004, 117, 3221–3231. [Google Scholar] [CrossRef] [Green Version]
- Di-Poï, N.; Fauré, J.; Grizot, S.; Molnár, G.; Pick, E.; Dagher, M.-C. Mechanism of NADPH oxidase activation by the Rac/Rho-GDI complex †. Biochemistry 2001, 40, 10014–10022. [Google Scholar] [CrossRef]
- DerMardirossian, C.; Schnelzer, A.; Bokoch, G.M. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell 2004, 15, 117–127. [Google Scholar] [CrossRef]
- DerMardirossian, C.; Bokoch, G.M. GDIs: Central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005, 15, 356–363. [Google Scholar] [CrossRef]
- Wiegandt, D.; Vieweg, S.; Hofmann, F.; Koch, D.; Li, F.; Wu, Y.-W.; Itzen, A.; Müller, M.P.; Goody, R.S. Locking GTPases covalently in their functional states. Nat. Commun. 2015, 6, 7773. [Google Scholar] [CrossRef] [Green Version]
- Akula, M.K.; Ibrahim, M.X.; Ivarsson, E.G.; Khan, O.M.; Kumar, I.T.; Erlandsson, M.; Karlsson, C.; Xu, X.; Brisslert, M.; Brakebusch, C.; et al. Protein prenylation restrains innate immunity by inhibiting Rac1 effector interactions. Nat. Commun. 2019, 10, 3975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozza, W.P.; Zhang, Y.; Hallett, K.; Rosado, L.A.R.; Zhang, B. RhoGDI deficiency induces constitutive activation of Rho GTPases and COX-2 pathways in association with breast cancer progression. Oncotarget 2015, 6, 32723–32736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dovas, A.; Couchman, J.R. RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Lérida, I.; Pellinen, T.; Sanchez, S.A.; Guadamillas, M.C.; Wang, Y.; Mirtti, T.; Calvo, E.; Del Pozo, M.A. Rac1 nucleocytoplasmic shuttling drives nuclear shape changes and tumor invasion. Dev. Cell 2015, 32, 318–334. [Google Scholar] [CrossRef] [Green Version]
- Michaelson, D.; Abidi, W.; Guardavaccaro, D.; Zhou, M.; Ahearn, I.; Pagano, M.; Philips, M.R. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J. Cell Biol. 2008, 181, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Concepcion, M. The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 1999, 18, 1996–2007. [Google Scholar] [CrossRef] [Green Version]
- Hofemeister, H.; Weber, K.; Stick, R. Association of prenylated proteins with the plasma membrane and the inner nuclear membrane is mediated by the same membrane-targeting motifs. Mol. Biol. Cell 2000, 11, 3233–3246. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Zheng, Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin. Drug Discov. 2015, 10, 991–1010. [Google Scholar] [CrossRef] [Green Version]
- Cardama, G.A.; Gonzalez, N.; Maggio, J.; Menna, P.L.; Gomez, D.E. Rho GTPases as therapeutic targets in cancer (Review). Int. J. Oncol. 2017, 51, 1025–1034. [Google Scholar] [CrossRef] [Green Version]
- Clayton, N.S.; Ridley, A.J. Targeting Rho GTPase signaling networks in cancer. Front. Cell Dev. Biol. 2020, 8, 222. [Google Scholar] [CrossRef]
- Sarrabayrouse, G.; Pich, C.; Teiti, I.; Tilkin-Mariame, A.F. Regulatory properties of statins and Rho GTPases prenylation inhibitors to stimulate melanoma immunogenicity and promote anti-melanoma immune response. Int. J. Cancer 2017, 140, 747–755. [Google Scholar] [CrossRef]
- Abdullah, M.I.; de Wolf, E.; Jawad, M.J.; Richardson, A. The poor design of clinical trials of statins in oncology may explain their failure—Lessons for drug repurposing. Cancer Treat. Rev. 2018, 69, 84–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aicart-Ramos, C.; Valero, R.A.; Rodriguez-Crespo, I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 2981–2994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greaves, J.; Chamberlain, L.H. Palmitoylation-dependent protein sorting. J. Cell Biol. 2007, 176, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Won, S.J.; Martin, B.R. Temporal profiling establishes a dynamic S-palmitoylation cycle. ACS Chem. Biol. 2018, 13, 1560–1568. [Google Scholar] [CrossRef]
- Navarro-Lérida, I.; Álvarez-Barrientos, A.; Gavilanes, F.; Rodríguez-Crespo, I. Distance-dependent cellular palmitoylation of de-novo-designed sequences and their translocation to plasma membrane subdomains. J. Cell Sci. 2002, 115, 3119–3130. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhi, X.; Wang, X.; Meng, D. Protein palmitoylation and its pathophysiological relevance. J. Cell. Physiol. 2020, 5, 3220–3233. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, R.; Fukata, Y.; Fukata, M. Discovery of protein-palmitoylating enzymes. Pflüg. Arch. Eur. J. Physiol. 2008, 456, 1199–1206. [Google Scholar] [CrossRef]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.-P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Tabaczar, S.; Czogalla, A.; Podkalicka, J.; Biernatowska, A.; Sikorski, A.F. Protein palmitoylation: Palmitoyltransferases and their specificity. Exp. Biol. Med. 2017, 242, 1150–1157. [Google Scholar] [CrossRef] [Green Version]
- Garland, M.; Schulze, C.J.; Foe, I.T.; van der Linden, W.A.; Child, M.A.; Bogyo, M. Development of an activity-based probe for acyl-protein thioesterases. PLoS ONE 2018, 13, e0190255. [Google Scholar] [CrossRef] [Green Version]
- Rusch, M.; Zimmermann, T.J.; Bürger, M.; Dekker, F.J.; Görmer, K.; Triola, G.; Brockmeyer, A.; Janning, P.; Böttcher, T.; Sieber, S.A.; et al. Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew. Chem. Int. Ed. 2011, 50, 9838–9842. [Google Scholar] [CrossRef]
- Hancock, J.F.; Magee, A.I.; Childs, J.E.; Marshall, C.J. All Ras proteins are polyisoprenylated but only some are palmitoylated. Cell 1989, 57, 1167–1177. [Google Scholar] [CrossRef]
- Laude, A.J.; Prior, I.A. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J. Cell Sci. 2008, 121, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Lynch, S.J.; Snitkin, H.; Gumper, I.; Philips, M.R.; Sabatini, D.; Pellicer, A. The differential palmitoylation states of N-Ras and H-Ras determine their distinct Golgi subcompartment localizations. J. Cell. Physiol. 2015, 230, 610–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamson, P.; Paterson, H.E.; Hall, A. Intracellular localization of the p21 Rho proteins. J. Cell Biol. 1992, 119, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.-A.; Sebti, S.M. Palmitoylated cysteine 192 is required for RhoB tumor-suppressive and apoptotic activities. J. Biol. Chem. 2005, 280, 19243–19249. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Lérida, I.; Sánchez-Perales, S.; Calvo, M.; Rentero, C.; Zheng, Y.; Enrich, C.; del Pozo, M.A. A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 2012, 31, 534–551. [Google Scholar] [CrossRef] [Green Version]
- Kang, R.; Wan, J.; Arstikaitis, P.; Takahashi, H.; Huang, K.; Bailey, A.O.; Thompson, J.X.; Roth, A.F.; Drisdel, R.C.; Mastro, R.; et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 2008, 456, 904–909. [Google Scholar] [CrossRef] [Green Version]
- Albanesi, J.P.; Barylko, B.; DeMartino, G.N.; Jameson, D.M. Palmitoylated proteins in dendritic spine remodeling. Front. Synaptic Neurosci. 2020, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Druso, J.E.; Cerione, R.A. The two splice variant forms of Cdc42 exert distinct and essential functions in neurogenesis. J. Biol. Chem. 2020, 295, 4498–4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Borja, M. RhoB regulates endosome transport by promoting actin assembly on endosomal membranes through Dia1. J. Cell Sci. 2005, 118, 2661–2670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valero, R.A.; Oeste, C.L.; Stamatakis, K.; Ramos, I.; Herrera, M.; Boya, P.; Pérez-Sala, D. Structural determinants allowing endolysosomal sorting and degradation of endosomal GTPases. Traffic 2010, 11, 1221–1233. [Google Scholar] [CrossRef] [Green Version]
- Strehle, A.; Schleicher, M.; Faix, J. Trix, a novel Rac guanine-nucleotide exchange factor from Dictyostelium discoideum is an actin-binding protein and accumulates at endosomes. Eur. J. Cell Biol. 2006, 85, 1035–1045. [Google Scholar] [CrossRef]
- Palamidessi, A.; Frittoli, E.; Garré, M.; Faretta, M.; Mione, M.; Testa, I.; Diaspro, A.; Lanzetti, L.; Scita, G.; di Fiore, P.P. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell 2008, 134, 135–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phuyal, S.; Farhan, H. Multifaceted Rho GTPase signaling at the endomembranes. Front. Cell Dev. Biol. 2019, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Osmani, N.; Peglion, F.; Chavrier, P.; Etienne-Manneville, S. Cdc42 localization and cell polarity depend on membrane traffic. J. Cell Biol. 2010, 191, 1261–1269. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, T. Emerging roles of recycling endosomes. J. Biochem. 2013, 153, 505–510. [Google Scholar] [CrossRef] [Green Version]
- Kirkham, M.; Parton, R.G. Clathrin-independent endocytosis: New insights into caveolae and non-caveolar lipid raft carriers. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2005, 1745, 273–286. [Google Scholar] [CrossRef] [Green Version]
- Misaki, R.; Morimatsu, M.; Uemura, T.; Waguri, S.; Miyoshi, E.; Taniguchi, N.; Matsuda, M.; Taguchi, T. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J. Cell Biol. 2010, 191, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Bekhouche, B.; Tourville, A.; Ravichandran, Y.; Tacine, R.; Abrami, L.; Dussiot, M.; Khau-Dancasius, A.; Boccara, O.; Khirat, M.; Mangeney, M.; et al. A toxic palmitoylation of Cdc42 enhances NF-ΚB signaling and drives a severe autoinflammatory syndrome. J. Allergy Clin. Immunol. 2020, 146, 1201–1204. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Yin, T.; Yang, Q.; Zhang, J.; Wu, Y.I.; Yu, J. Single-molecule tracking of small GTPase Rac1 uncovers spatial regulation of membrane translocation and mechanism for polarized signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remorino, A.; de Beco, S.; Cayrac, F.; di Federico, F.; Cornilleau, G.; Gautreau, A.; Parrini, M.C.; Masson, J.-B.; Dahan, M.; Coppey, M. Gradients of Rac1 nanoclusters support spatial patterns of Rac1 signaling. Cell Rep. 2017, 21, 1922–2935. [Google Scholar] [CrossRef] [Green Version]
- Moissoglu, K.; Kiessling, V.; Wan, C.; Hoffman, B.D.; Norambuena, A.; Tamm, L.K.; Schwartz, M.A. Regulation of Rac1 translocation and activation by membrane domains and their boundaries. J. Cell Sci. 2014, 127, 2565–2576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, K.N.; Zhou, Y.; Hancock, J.F. Rac1 nanoscale organization on the plasma membrane is driven by lipid binding specificity encoded in the membrane anchor. Mol. Cell. Biol. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Saliba, A.-E.; Vonkova, I.; Ceschia, S.; Findlay, G.M.; Maeda, K.; Tischer, C.; Deghou, S.; van Noort, V.; Bork, P.; Pawson, T.; et al. A Quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat. Methods 2014, 11, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Bakal, C.; Aach, J.; Church, G.; Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 2007, 316, 1753–1756. [Google Scholar] [CrossRef] [Green Version]
- Crisp, M.; Burke, B. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 2008, 582, 2023–2031. [Google Scholar] [CrossRef] [Green Version]
- Grossman, E.; Medalia, O.; Zwerger, M. Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 2012, 41, 557–584. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.; Fusco, S.; Grassi, C. Nutrient-dependent changes of protein palmitoylation: Impact on nuclear enzymes and regulation of gene expression. Int. J. Mol. Sci. 2018, 19, 3820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinelli, M.; Fusco, S.; Mainardi, M.; Scala, F.; Natale, F.; Lapenta, R.; Mattera, A.; Rinaudo, M.; Li Puma, D.D.; Ripoli, C.; et al. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through FoxO3a. Nat. Commun. 2017, 8, 2009. [Google Scholar] [CrossRef]
- Pedram, A.; Razandi, M.; Sainson, R.C.A.; Kim, J.K.; Hughes, C.C.; Levin, E.R. A conserved mechanism for steroid receptor translocation to the plasma membrane. J. Biol. Chem. 2007, 282, 22278–22288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhaber, B.; Sammer, M.; Lua, W.H.; Benetka, W.; Liew, L.L.; Yu, W.; Lee, H.K.; Koranda, M.; Eisenhaber, F.; Adhikari, S. Nuclear import of a lipid-modified transcription factor. Cell Cycle 2011, 10, 3897–3911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, K.F.; Erickson, J.W.; Antonyak, M.A.; Cerione, R.A. Rho GTPases and their roles in cancer metabolism. Trends Mol. Med. 2013, 19, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Baidwan, S.; Chekuri, A.; Hynds, D.L.; Kowluru, A. Glucotoxicity promotes aberrant activation and mislocalization of Ras-related C3 botulinum toxin substrate 1 [Rac1] and metabolic dysfunction in pancreatic islet β-cells: Reversal of such metabolic defects by metformin. Apoptosis 2017, 22, 1380–1393. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.; Reinhard, N.R.; Hordijk, P.L. Toward understanding RhoGTPase specificity: Structure, function and local activation. Small GTPases 2014, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Lanning, C.C.; Daddona, J.L.; Ruiz-Velasco, R.; Shafer, S.H.; Williams, C.L. The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J. Biol. Chem. 2004, 279, 44197–44210. [Google Scholar] [CrossRef] [Green Version]
- Dubash, A.D.; Guilluy, C.; Srougi, M.C.; Boulter, E.; Burridge, K.; García-Mata, R. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS ONE 2011, 6, e17380. [Google Scholar] [CrossRef] [Green Version]
- Abdrabou, A.; Wang, Z. Regulation of the nuclear speckle localization and function of Rac1. FASEB J. 2021, 35, e21235. [Google Scholar] [CrossRef]
- Williams, C.L. The polybasic region of Ras and Rho family small GTPases: A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell. Signal. 2003, 15, 1071–1080. [Google Scholar] [CrossRef]
- Sandrock, K.; Bielek, H.; Schradi, K.; Schmidt, G.; Klugbauer, N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin A2. Traffic 2010, 11, 198–209. [Google Scholar] [CrossRef]
- Casado-Medrano, V.; Barrio-Real, L.; Gutiérrez-Miranda, L.; González-Sarmiento, R.; Velasco, E.A.; Kazanietz, M.G.; Caloca, M.J. Identification of a truncated Β1-chimaerin variant that inactivates nuclear Rac1. J. Biol. Chem. 2020, 295, 1300–1314. [Google Scholar] [CrossRef]
- Kelpsch, D.J.; Tootle, T.L. Nuclear actin: From discovery to function. Anat. Rec. 2018, 301, 1999–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jockusch, B.M.; Schoenenberger, C.-A.; Stetefeld, J.; Aebi, U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006, 16, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Baarlink, C.; Wang, H.; Grosse, R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013, 340, 864–867. [Google Scholar] [CrossRef]
- Baarlink, C.; Plessner, M.; Sherrard, A.; Morita, K.; Misu, S.; Virant, D.; Kleinschnitz, E.-M.; Harniman, R.; Alibhai, D.; Baumeister, S.; et al. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 2017, 19, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Plessner, M.; Grosse, R. Extracellular signaling cues for nuclear actin polymerization. Eur. J. Cell Biol. 2015, 94, 359–362. [Google Scholar] [CrossRef]
- Rottner, K.; Faix, J.; Bogdan, S.; Linder, S.; Kerkhoff, E. Actin assembly mechanisms at a glance. J. Cell Sci. 2017, 130, 3427–3435. [Google Scholar] [CrossRef] [Green Version]
- Weston, L.; Coutts, A.S.; la Thangue, N.B. Actin nucleators in the nucleus: An emerging theme. J. Cell Sci. 2012, 125, 3519–3527. [Google Scholar] [CrossRef] [Green Version]
- Fiore, A.P.Z.P.; Spencer, V.A.; Mori, H.; Carvalho, H.F.; Bissell, M.J.; Bruni-Cardoso, A. Laminin-111 and the level of nuclear actin regulate epithelial quiescence via exportin-6. Cell Rep. 2017, 19, 2102–2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurica, M.S.; Licklider, L.J.; Gygi, S.P.; Grigorieff, N.; Moore, M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 2002, 8, 426–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitoh, N.; Spahr, C.S.; Patterson, S.D.; Bubulya, P.; Neuwald, A.F.; Spector, D.L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 2004, 15, e03–e04. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parisis, N.; Krasinska, L.; Harker, B.; Urbach, S.; Rossignol, M.; Camasses, A.; Dewar, J.; Morin, N.; Fisher, D. Initiation of DNA replication requires actin dynamics and formin activity. EMBO J. 2017, 36, 3212–3231. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Hu, Y.; Belmont, A.S. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 2014, 24, 1138–1144. [Google Scholar] [CrossRef] [Green Version]
- Maniotis, A.J.; Chen, C.S.; Ingber, D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA 1997, 94, 849–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear lamins: Thin filaments with major functions. Trends Cell Biol. 2018, 28, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Sasseville, A.M.-J.; Langelier, Y. In vitro interaction of the carboxy-terminal domain of Lamin A with actin. FEBS Lett. 1998, 425, 485–489. [Google Scholar] [CrossRef]
- Simon, D.N.; Zastrow, M.S.; Wilson, K.L. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 2010, 1, 264–272. [Google Scholar] [CrossRef]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef]
- Percipalle, P.; Vartiainen, M. Cytoskeletal proteins in the cell nucleus: A special nuclear actin perspective. Mol. Biol. Cell 2019, 30, 1781–1785. [Google Scholar] [CrossRef]
- Fritz, G.; Henninger, C. Rho GTPases: Novel players in the regulation of the DNA damage response? Biomolecules 2015, 5, 2417–2434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, M.L.; Lammerding, J. Keeping the LINC: The importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 2011, 39, 1729–1734. [Google Scholar] [CrossRef] [Green Version]
- Kirby, T.J.; Lammerding, J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 2018, 20, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K.; Monaghan-Benson, E.; Graham, D.M. Mechanotransduction: From the cell surface to the nucleus via RhoA. Philos. Trans. R. Soc. B: Biol. Sci. 2019, 374, 20180229. [Google Scholar] [CrossRef] [PubMed]
- Colón-Bolea, P.; García-Gómez, R.; Shackleton, S.; Crespo, P.; Bustelo, X.R.; Casar, B. Rac1 induces nuclear alterations through the LINC complex to enhance melanoma invasiveness. Mol. Biol. Cell 2020, 31, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Woroniuk, A.; Porter, A.; White, G.; Newman, D.T.; Diamantopoulou, Z.; Waring, T.; Rooney, C.; Strathdee, D.; Marston, D.J.; Hahn, K.M.; et al. STEF/TIAM2-mediated Rac1 activity at the nuclear envelope regulates the perinuclear actin cap. Nat. Commun. 2018, 9, 2124. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017, 216, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.; Barton, M.; Nguyen, N. Stretching induces overexpression of RhoA and Rac1 GTPases in breast cancer cells. Adv. Biosyst. 2020, 4. [Google Scholar] [CrossRef] [PubMed]
- Kunschmann, T.; Puder, S.; Fischer, T.; Steffen, A.; Rottner, K.; Mierke, C.T. The small GTPase Rac1 increases cell surface stiffness and enhances 3D migration into extracellular matrices. Sci. Rep. 2019, 9, 7675. [Google Scholar] [CrossRef] [PubMed]
- Donnaloja, F.; Jacchetti, E.; Soncini, M.; Raimondi, M.T. Mechanosensing at the nuclear envelope by nuclear pore complex stretch activation and its effect in physiology and pathology. Front. Physiol. 2019, 10, 896. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, W.K.; Tam, A.; Fujimura-Kamada, K.; Michaelis, S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 11175–11180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampton, S.E.; Dore, T.M.; Schmidt, W.K. Rce1: Mechanism and inhibition. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 157–174. [Google Scholar] [CrossRef]
- Lemmer, I.L.; Willemsen, N.; Hilal, N.; Bartelt, A. A guide to understanding endoplasmic reticulum stress in metabolic disorders. Mol. Metab. 2021, 47. [Google Scholar] [CrossRef]
- Philippe, J.M.; Jenkins, P.M. Spatial organization of palmitoyl acyl transferases governs substrate localization and function. Mol. Membr. Biol. 2019, 35, 60–75. [Google Scholar] [CrossRef]
- Gorleku, O.A.; Barns, A.-M.; Prescott, G.R.; Greaves, J.; Chamberlain, L.H. Endoplasmic reticulum localization of DHHC palmitoyltransferases mediated by lysine-based sorting signals. J. Biol. Chem. 2011, 286, 39573–39584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chrysanthou, E.; Gorringe, K.L.; Joseph, C.; Craze, M.; Nolan, C.C.; Diez-Rodriguez, M.; Green, A.R.; Rakha, E.A.; Ellis, I.O.; Mukherjee, A. Phenotypic characterisation of breast cancer: The role of CDC42. Breast Cancer Res. Treat. 2017, 164, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.S.; Drubin, D.G. Cdc42 GTPase regulates ESCRTs in nuclear envelope sealing and ER remodeling. J. Cell Biol. 2020, 219, e201910119. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, J.; Liang, X.; Yin, X.; Zhang, L.; Chen, D.; Jin, X.; Fiskesund, R.; Tang, K.; Ma, J.; et al. Fibrin stiffness mediates dormancy of tumor-repopulating cells via a Cdc42-driven Tet2 epigenetic program. Cancer Res. 2018, 78, 3926–3937. [Google Scholar] [CrossRef] [Green Version]
- Hunt, A.N. Dynamic Lipidomics of the nucleus. J. Cell. Biochem. 2006, 97, 244–251. [Google Scholar] [CrossRef]
- Albi, E.; Magni, M.P.V. The role of intranuclear lipids. Biol. Cell 2004, 96, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Radhakrishnan, A.; Goldstein, J.L. Retrospective on cholesterol homeostasis: The central role of SCAP. Annu. Rev. Biochem. 2018, 87, 783–807. [Google Scholar] [CrossRef] [Green Version]
- Dazzoni, R.; Grélard, A.; Morvan, E.; Bouter, A.; Applebee, C.J.; Loquet, A.; Larijani, B.; Dufourc, E.J. The unprecedented membrane deformation of the human nuclear envelope, in a magnetic field, indicates formation of nuclear membrane invaginations. Sci. Rep. 2020, 10, 5147. [Google Scholar] [CrossRef] [Green Version]
- Saxena, N.; Chandra, N.C. Cholesterol: A prelate in cell nucleus and its serendipity. Curr. Mol. Med. 2021, 20, 692–707. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy-Kanniappan, S. Rac1 repression reverses chemoresistance by targeting tumor metabolism. Cancer Biol. Ther. 2020, 21, 888–890. [Google Scholar] [CrossRef]
- Schnelzer, A.; Prechtel, D.; Knaus, U.; Dehne, K.; Gerhard, M.; Graeff, H.; Harbeck, N.; Schmitt, M.; Lengyel, E. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000, 19, 3013–3020. [Google Scholar] [CrossRef] [Green Version]
- Goka, E.T.; Chaturvedi, P.; Lopez, D.T.M.; de La Garza, A.; Lippman, M.E. RAC1b overexpression confers resistance to chemotherapy treatment in colorectal cancer. Mol. Cancer Ther. 2019, 18, 957–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, C.; Hass, R.; Lehnert, H.; Ungefroren, H. Rac1B: A Rho GTPase with versatile functions in malignant transformation and tumor progression. Cells 2019, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Seiz, J.R.; Klinke, J.; Scharlibbe, L.; Lohfink, D.; Heipel, M.; Ungefroren, H.; Giehl, K.; Menke, A. Different signaling and functionality of Rac1 and Rac1b in the progression of lung adenocarcinoma. Biol. Chem. 2020, 401, 517–531. [Google Scholar] [CrossRef]
- Vu, H.L.; Rosenbaum, S.; Purwin, T.J.; Davies, M.A.; Aplin, A.E. Rac1 P29S regulates PD-L1 expression in melanoma. Pigment. Cell Melanoma Res. 2015, 28, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Revach, O.-Y.; Winograd-Katz, S.E.; Samuels, Y.; Geiger, B. The involvement of mutant Rac1 in the formation of invadopodia in cultured melanoma cells. Exp. Cell Res. 2016, 343, 82–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, P.; Aske, J.C.; Dey, N. RAC1 Takes the lead in solid tumors. Cells 2019, 8, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| RhoGTPases | Post-Translational Modifications |
|---|---|
| Rac1 | Prenylation Cys189 Palmitoylation Cys178 Phosphorylation: Tyr32; Tyr64; Ser71; Thr108 Ubiquitylation: Lys147, Lys166 Sumoylation: PBR domain Glutathionylation Cys18, Cys81, Cys118, Cys157 |
| Rac2 | Prenylation Cys189 S-Glutathionylation Cys157 |
| Rac3 | Prenylation Cys189 Ubiquitylation Lys166 |
| Cdc42 | Prenylation Cys189 Palmitoylation Cys188 Phosphorylation: Tyr32; Tyr64; Ser71 Ubiquitylation: Lys147, Lys166 |
| RhoA | Prenylation Cys190 Phosphorylation: Ser26, Tyr32, Tyr66, Ser188 Ubiquitylation: Lys6, Lys7, Lys135 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro-Lérida, I.; Sánchez-Álvarez, M.; del Pozo, M.Á. Post-Translational Modification and Subcellular Compartmentalization: Emerging Concepts on the Regulation and Physiopathological Relevance of RhoGTPases. Cells 2021, 10, 1990. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10081990
Navarro-Lérida I, Sánchez-Álvarez M, del Pozo MÁ. Post-Translational Modification and Subcellular Compartmentalization: Emerging Concepts on the Regulation and Physiopathological Relevance of RhoGTPases. Cells. 2021; 10(8):1990. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10081990
Chicago/Turabian StyleNavarro-Lérida, Inmaculada, Miguel Sánchez-Álvarez, and Miguel Ángel del Pozo. 2021. "Post-Translational Modification and Subcellular Compartmentalization: Emerging Concepts on the Regulation and Physiopathological Relevance of RhoGTPases" Cells 10, no. 8: 1990. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10081990