Molecular Characterization of NDL1-AGB1 Mediated Salt Stress Signaling: Further Exploration of the Role of NDL1 Interacting Partners
Abstract
:1. Introduction
2. Material and Methods
2.1. In-Silico Analysis
2.2. Phenotypic Analysis under Salt Stress
2.3. Cloning
2.4. Yeast Complementation Assay
2.5. Yeast Salt Stress-Tolerance Growth Assay
3. Results
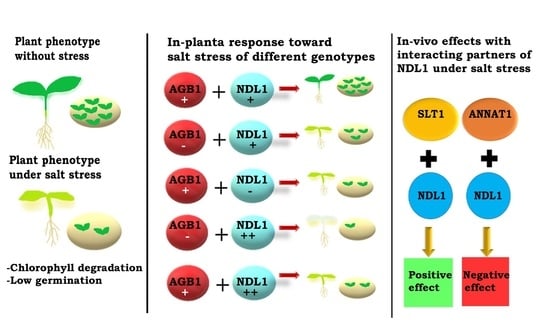
3.1. NDL1 Acts as Negative Regulator of Rosette Diameter and Chlorophyll Leaching and Work Downstream of AGB1 during Salt Stress
3.2. NDL1 Acts as Positive Regulator of Germination during Salinity Stress and Work Downstream of AGB1
3.3. Salt Response Specific Components of NDL1 Interactome–Expression during Different Developmental Stages and Tissues
3.4. Detailed In-Silico Expression Analysis of SRPIN under Salt Stress
3.5. Complementation and Stress Assays in Yeast to Confirm In Vivo Interactions between NDL1 and Select Candidates of SRPIN and Confirm Their Functional Dependency
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhu, J.K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [Green Version]
- Conde, A.; Chaves, M.M.; Gerós, H. Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol. 2011, 52, 1583–1602. [Google Scholar] [CrossRef]
- Maathuis, F.J. Sodium in plants: Perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 2014, 65, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.G.; Jones, A.M. AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 2004, 389, 338–350. [Google Scholar] [CrossRef]
- Chen, J.G.; Willard, F.S.; Huang, J.; Liang, J.; Chasse, S.A.; Jones, A.M.; Siderovski, D.P. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 2003, 301, 1728–1731. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.Y.; Urano, D. Genetic and systematic approaches toward G protein-coupled abiotic stress signaling in plants. Front. Plant Sci. 2018, 9, 1378. [Google Scholar] [CrossRef] [PubMed]
- Colaneri, A.C.; Tunc-Ozdemir, M.; Huang, J.P.; Jones, A.M. Growth attenuation under saline stress is mediated by the heterotrimeric G protein complex. BMC Plant Biol. 2014, 14, 129. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Assmann, S.M. The heterotrimeric G-protein β subunit, AGB 1, plays multiple roles in the Arabidopsis salinity response. Plant Cell Environ. 2015, 38, 2143–2156. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gao, Y.; Jones, A.M. Extra Large G-Protein Interactome Reveals Multiple Stress Response Function and Partner-Dependent XLG Subcellular Localization. Front. Plant Sci. 2017, 8, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urano, D.; Colaneri, A.; Jones, A.M. Gα modulates salt-induced cellular senescence and cell division in rice and maize. J. Exp. Bot. 2014, 65, 6553–6561. [Google Scholar] [CrossRef] [Green Version]
- Swain, D.M.; Sahoo, R.K.; Srivastava, V.K.; Tripathy, B.C.; Tuteja, R.; Tuteja, N. Function of heterotrimeric G-protein γ subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta 2017, 245, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Friedman, E.J.; Wang, H.X.; Jiang, K.; Perovic, I.; Deshpande, A.; Pochapsky, T.C.; Temple, B.R.; Hicks, S.N.; Harden, T.K.; Jones, A.M. Acireductonedioxygenase 1(ARD1) is an effector of the heterotrimeric G protein beta subunit in Arabidopsis. J. Biol. Chem. 2011, 286, 30107–30118. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Taylor, J.P.; Chen, J.G.; Uhrig, J.F.; Schnell, D.J.; Nakagawa, T.; Korth, K.L.; Jones, A.M. The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 2006, 18, 1226–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudgil, Y.; Uhrig, J.F.; Zhou, J.; Temple, B.; Jiang, K.; Jones, A.M. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell 2009, 21, 3591–3609. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.X.; Weerasinghe, R.R.; Perdue, T.D.; Cakmakci, N.G.; Taylor, J.P.; Marzluff, W.F.; Jones, A.M. A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol. Biol. Cell 2006, 10, 4257–4269. [Google Scholar] [CrossRef] [Green Version]
- Klopffleisch, K.; Phan, N.; Augustin, K.; Bayne, R.S.; Booker, K.S.; Botella, J.R.; Carpita, N.C.; Carr, T.; Chen, J.G.; Cooke, T.R.; et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 2011, 7, 532. [Google Scholar] [CrossRef]
- Kanojia, A.; Mudgil, M. Detailed InSilico Analysis of Arabidopsis N-mycDownregulatedLike (NDL) Interactome with Reference to Stress Sensing. Phytomorphology 2020, 70, 87–104. [Google Scholar]
- Krauter, C.R.; Roberte, B.; Jean-Luc, E.; Günther, H.; Wolfgang, F.; André, S. A transmitting tissue- and pollen-expressed protein from sunflower with sequence similarity to the human RTP protein. Plant Sci. 1997, 129, 191–202. [Google Scholar] [CrossRef]
- Salnikow, K.; Kluz, T.; Costa, M.; Piquemal, D.; Demidenko, Z.N.; Xie, K.; Blagosklonny, M.V. The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol. Cell. Biol. 2002, 22, 1734–1741. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Zhau, H.E.; Huang, W.C.; Iqbal, S.; Habib, F.K.; Sartor, O.; Cvitanovic, L.; Marshall, F.F.; Xu, Z.; Chung, L.W. cAMP-responsive element-binding protein regulates vascular endothelial growth factor expression: Implication in human prostate cancer bone metastasis. Oncogene 2007, 26, 5070–5077. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.F.; Lu, J.; Zou, Y.S.; Soh-Won, J.; Cohen, D.M.; Buttrick, P.M.; Cooper, D.R.; Steinberg, S.F.; Mackman, N.; Pinsky, D.J.; et al. Hypoxia-associated induction of early growth response-1 gene expression. J. Biol. Chem. 1999, 274, 15030–15040. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Salnikow, K.; Costa, M. Cap43, a novel gene specifically induced by Ni2+compounds. Cancer Res. 1998, 58, 2182–2189. [Google Scholar] [PubMed]
- Rymaszewski, W.; Vile, D.; Bediee, A.; Dauzat, M.; Luchaire, N.; Kamrowska, D.; Granier, C.; Hennig, J. Stress-Related Gene Expression Reflects Morphophysiological Responses to Water Deficit. Plant Physiol. 2017, 174, 1913–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, C.; Xu, D.; Fang, G.; Wang, E.; Gao, S.; Xu, Z.; Li, L.; Zhang, X.; Min, D. G-protein β subunit AGB1 positively regulates salt stress tolerance in Arabidopsis. J. Integr. Agric. 2015, 14, 314–325. [Google Scholar] [CrossRef]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [Green Version]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [PubMed]
- Fuxman Bass, J.I.; Reece-Hoyes, J.S.; Walhout, A.J. Colony Lift Colorimetric Assay for β-Galactosidase Activity. Cold Spring Harb. Protoc. 2016, 12, pdb-rot088963. [Google Scholar] [CrossRef] [Green Version]
- Cantero, A.; Barthakur, S.; Bushart, T.J.; Chou, S.; Morgan, R.O.; Fernandez, M.P. Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant Physiol. Biochem. 2006, 44, 13–24. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, K.J.; Kim, J.Y.; Kim, Y.O.; Kang, H. Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant Cell Physiol. 2007, 48, 221–231. [Google Scholar] [CrossRef]
- Barroso, C.; Romero, L.C.; Cejudo, F.J.; Vega, J.M.; Gotor, C. Salt-specific regulation of thecytosolic O-acetylserine(thiol)lyase gene from Arabidopsis thaliana is dependentonabscisic acid. Plant Mol. Biol. 1999, 40, 729–736. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Arisz, S.A.; Dekker, H.L.; Kramer, G.; de Koster, C.G.; Haring, M.A. Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 2013, 450, 573–581. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, T.K.; Pardo, J.M.; Takeda, S.; Bressan, R.A.; Hasegawa, P.M. Tobacco and Arabidiopsis SLT1 mediate salt tolerance of yeast. Plant Mol. Biol. 2001, 45, 489–500. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular andmolecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Assmann, S.M. The effect of NaCl on stomatal opening in Arabidopsis wildtype and agb1 heterotrimeric G-protein mutant plants. Plant Signal Behav. 2016, 11, e1085275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katiyar, A.; Mudgil, Y. Play Role in Abiotic Stress and Hormonal Responses along with Their Specific Functions. Int. J. Mol. Sci. 2019, 20, 4736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Solís, J.R.; Gutierrez-Alcalá, G.; Vega, J.M.; Romero, L.C.; Gotor, C. Thecytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. J. Biol. Chem. 2001, 276, 9297–9302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatri, N.; Mudgil, Y. Hypothesis: NDL proteins function in stress responses by regulating microtubule organization. Front. Plant Sci. 2015, 6, 947. [Google Scholar] [CrossRef] [Green Version]
- Assmann, S.M. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 2002, 14 (Suppl. 1), S355–S373. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Chen, J.G.; Temple, B.; Boyes, D.C.; Alonso, J.M.; Davis, K.R.; Ecker, J.R.; Jones, A.M. The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 2003, 15, 393–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Chen, J.G.; Jones, A.M.; Assmann, S.M. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and post germination development. Plant Physiol. 2006, 141, 243–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, S.M.; Noh, E.K.; Kim, H.G.; Jeon, B.W.; Bae, K.; Hu, H.C.; Kwak, J.M.; Park, O.K. Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses. Plant Cell Physiol. 2010, 51, 1499–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| S.No. | Gene Name | Gene ID | Primers (5′ to 3′) |
|---|---|---|---|
| 1. | ANNAT1 | AT1G35720 | Forward CACCATGGCGACTCTTAAGGT |
| Reverse AGCATCATCTTCACC GAGAA | |||
| 2. | IDH-V | AT5G03290 | Forward CACCATGACCATGGCAGCAAA |
| Reverse GAGATGATCACAGATTGCCTTTG | |||
| 3. | SLT1 | AT2G37570 | Forward CACCATGGAGAATCATCATCCTTCT |
| Reverse TTAAGTCAGCATAAGATCGTTTCC |
| S.No. | Genes | Based on Online Available Sources | In-Silico above Two-Fold | References |
|---|---|---|---|---|
| 1 | AGB1 | √ | [27] | |
| 2 | ANNAT1 | √ | √ | [32] |
| 3 | ABHSP | √ | ||
| 4 | TIR900 | √ | ||
| 5 | TIR920 | √ | ||
| 6 | HP | √ | ||
| 7 | CYT4 | √ | ||
| 8 | NUCLEASE | √ | ||
| 9 | PEARLI4 | √ | ||
| 10 | PLCL | √ | ||
| 11 | VQ32 | √ | ||
| 12 | BOB1 | √ | ||
| 13 | CA1 | √ | ||
| 14 | CAD9 | √ | ||
| 15 | CKS2 | unpublished | ||
| 16 | COB | √ | [33] | |
| 17 | HMGB3 | √ | [34] | |
| 18 | LOX2 | √ | ||
| 19 | MT2A | √ | ||
| 20 | OASA1 | √ | √ | [35] |
| 21 | P14 GAMMA 4 | √ | √ | [36] |
| 22 | SLT1 | √ | √ | [37] |
| 23 | XT1 | √ | ||
| 24 | RAD5 | √ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, N.; Kanojia, A.; Katiyar, A.; Mudgil, Y. Molecular Characterization of NDL1-AGB1 Mediated Salt Stress Signaling: Further Exploration of the Role of NDL1 Interacting Partners. Cells 2021, 10, 2261. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10092261
Gupta N, Kanojia A, Katiyar A, Mudgil Y. Molecular Characterization of NDL1-AGB1 Mediated Salt Stress Signaling: Further Exploration of the Role of NDL1 Interacting Partners. Cells. 2021; 10(9):2261. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10092261
Chicago/Turabian StyleGupta, Nidhi, Abhishek Kanojia, Arpana Katiyar, and Yashwanti Mudgil. 2021. "Molecular Characterization of NDL1-AGB1 Mediated Salt Stress Signaling: Further Exploration of the Role of NDL1 Interacting Partners" Cells 10, no. 9: 2261. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10092261