Host-Cell Type Dependent Features of Recombinant Human Aquaporin-4 Orthogonal Arrays of Particles—New Insights for Structural and Functional Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line
2.2. Transfection and Baculovirus-Based Gene Expression
2.3. Antibodies
2.4. Patient Sera
2.5. Immunofluorescence
2.5.1. AQP4 Immunofluorescence
2.5.2. AQP4-IgG Immunofluorescence
2.6. Confocal Imaging and Analysis
2.7. DDM/SDS Solubility Assay and Western Blotting
2.8. Blue Native-PAGE and 2DE
2.9. Preparation of Membrane Vesicles
2.10. Native Size Exclusion Chromatography
2.11. Dot Blot
2.12. Sandwich ELISA
2.13. Densitometric Analysis
2.14. Statistical Analysis
3. Results
3.1. Analysis of AQP4 Localization and OAP Assembly into the Cell Membrane in Insect and Mammalian Cell Lines
3.2. Analysis of AQP4 Folding in Insect and Mammalian Cell Lines
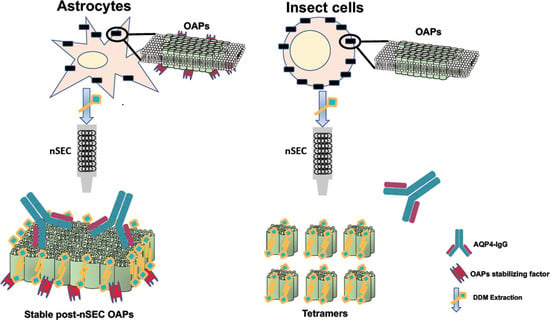
3.3. Analysis of OAPs Post-Extraction Integrity Performed by Blue-Native SDS/PAGE and Native Size Exclusion Chromatography (nSEC)
3.4. Evaluation of OAP Post-Extraction Stability by AQP4-IgGs Binding in nSEC Elutes and Plasma Membrane Vesicles Extracts
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Abbott, N.J.; Pizzo, M.E.; Preston, J.E.; Janigro, D.; Thorne, R.G. The role of brain barriers in fluid movement in the CNS: Is there a ‘glymphatic’ system? Acta Neuropathol. 2018, 135, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Nicchia, G.P.; Nico, B.; Camassa, L.M.; Mola, M.G.; Loh, N.; Dermietzel, R.; Spray, D.C.; Svelto, M.; Frigeri, A. The role of aquaporin-4 in the blood–brain barrier development and integrity: Studies in animal and cell culture models. Neuroscience 2004, 129, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Nico, B.; Frigeri, A.; Nicchia, G.P.; Quondamatteo, F.; Herken, R.; Errede, M.; Ribatti, D.; Svelto, M.; Roncali, L. Role of aquaporin-4 water channel in the development and integrity of the blood–brain barrier. J. Cell Sci. 2001, 114, 1297–1307. [Google Scholar] [PubMed]
- Frigeri, A.; Gropper, M.A.; Turck, C.W.; Verkman, A.S. Immunolocalization of the mercurial-insensitive water channel and glycerol intrinsic protein in epithelial cell plasma membranes. Proc. Natl. Acad. Sci. USA 1995, 92, 4328–4331. [Google Scholar] [CrossRef] [PubMed]
- De Bellis, M.; Pisani, F.; Mola, M.G.; Basco, D.; Catalano, F.; Nicchia, G.P.; Svelto, M.; Frigeri, A. A novel human aquaporin-4 splice variant exhibits a dominant-negative activity: A new mechanism to regulate water permeability. Mol. Biol. Cell 2014, 25, 470–480. [Google Scholar] [CrossRef]
- De Bellis, M.; Pisani, F.; Mola, M.G.; Rosito, S.; Simone, L.; Buccoliero, C.; Trojano, M.; Nicchia, G.P.; Svelto, M.; Frigeri, A. Translational readthrough generates new astrocyte AQP4 isoforms that modulate supramolecular clustering, glial endfeet localization, and water transport. Glia 2017, 65, 790–803. [Google Scholar] [CrossRef]
- Rossi, A.; Crane, J.M.; Verkman, A.S. Aquaporin-4 Mz isoform: Brain expression, supramolecular assembly and neuromyelitis optica antibody binding. Glia 2011, 59, 1056–1063. [Google Scholar] [CrossRef] [Green Version]
- Wolburg, H.; Berg-von der Emde, K.; Naujoks-Manteuffel, C. Muller (glial) cells in the retina of urodeles and anurans reveal different morphology by means of freeze-fracturing. Neurosci. Lett. 1992, 138, 89–92. [Google Scholar] [CrossRef]
- Wolburg, H. Orthogonal arrays of intramembranous particles: A review with special reference to astrocytes. J. Hirnforsch. 1995, 36, 239–258. [Google Scholar]
- Rossi, A.; Moritz, T.J.; Ratelade, J.; Verkman, A.S. Super-resolution imaging of aquaporin-4 orthogonal arrays of particles in cell membranes. J. Cell Sci. 2012, 125, 4405–4412. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.J.; Jin, B.J.; Ratelade, J.; Verkman, A.S. Aggregation state determines the localization and function of M1- and M23-aquaporin-4 in astrocytes. J. Cell Biol. 2014, 204, 559–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crane, J.M.; Tajima, M.; Verkman, A.S. Live-cell imaging of aquaporin-4 diffusion and interactions in orthogonal arrays of particles. Neuroscience 2010, 168, 892–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkman, A.S.; Rossi, A.; Crane, J.M. Live-cell imaging of aquaporin-4 supramolecular assembly and diffusion. Methods Enzymol. 2012, 504, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Verbavatz, J.M.; Ma, T.; Gobin, R.; Verkman, A.S. Absence of orthogonal arrays in kidney, brain and muscle from transgenic knockout mice lacking water channel aquaporin-4. J. Cell Sci. 1997, 110 Pt 22, 2855–2860. [Google Scholar]
- Frigeri, A.; Gropper, M.A.; Umenishi, F.; Kawashima, M.; Brown, D.; Verkman, A.S. Localization of MIWC and GLIP water channel homologs in neuromuscular, epithelial and glandular tissues. J. Cell Sci. 1995, 108 Pt 9, 2993–3002. [Google Scholar]
- Nicchia, G.P.; Mastrototaro, M.; Rossi, A.; Pisani, F.; Tortorella, C.; Ruggieri, M.; Lia, A.; Trojano, M.; Frigeri, A.; Svelto, M. Aquaporin-4 orthogonal arrays of particles are the target for neuromyelitis optica autoantibodies. Glia 2009, 57, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Pisani, F.; Nicchia, G.P.; Svelto, M.; Frigeri, A. Evidences for a leaky scanning mechanism for the synthesis of the shorter M23 protein isoform of aquaporin-4: Implication in orthogonal array formation and neuromyelitis optica antibody interaction. J. Biol. Chem. 2010, 285, 4562–4569. [Google Scholar] [CrossRef]
- Pisani, F.; Mastrototaro, M.; Rossi, A.; Nicchia, G.P.; Tortorella, C.; Ruggieri, M.; Trojano, M.; Frigeri, A.; Svelto, M. Identification of two major conformational aquaporin-4 epitopes for neuromyelitis optica autoantibody binding. J. Biol. Chem. 2011, 286, 9216–9224. [Google Scholar] [CrossRef]
- Pisani, F.; Mola, M.G.; Simone, L.; Rosito, S.; Alberga, D.; Mangiatordi, G.F.; Lattanzi, G.; Nicolotti, O.; Frigeri, A.; Svelto, M.; et al. Identification of a point mutation impairing the binding between aquaporin-4 and neuromyelitis optica autoantibodies. J. Biol. Chem. 2014, 289, 30578–30589. [Google Scholar] [CrossRef]
- Crane, J.M.; Verkman, A.S. Determinants of aquaporin-4 assembly in orthogonal arrays revealed by live-cell single-molecule fluorescence imaging. J. Cell Sci. 2009, 122, 813–821. [Google Scholar] [CrossRef] [Green Version]
- Jin, B.J.; Rossi, A.; Verkman, A.S. Model of aquaporin-4 supramolecular assembly in orthogonal arrays based on heterotetrameric association of M1-M23 isoforms. Biophys. J. 2011, 100, 2936–2945. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Baumgart, F.; van Hoek, A.N.; Verkman, A.S. Post-Golgi supramolecular assembly of aquaporin-4 in orthogonal arrays. Traffic 2012, 13, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Pisani, F.; Simone, L.; Gargano, C.D.; De Bellis, M.; Cibelli, A.; Mola, M.G.; Catacchio, G.; Frigeri, A.; Svelto, M.; Nicchia, G.P. Role of the H-bond between L53 and T56 for Aquaporin-4 epitope in Neuromyelitis Optica. Biochim. Biophys. Acta 2017, 1859, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Rosito, S.; Nicchia, G.P.; Palazzo, C.; Lia, A.; Buccoliero, C.; Pisani, F.; Svelto, M.; Trojano, M.; Frigeri, A. Supramolecular aggregation of aquaporin-4 is different in muscle and brain: Correlation with tissue susceptibility in neuromyelitis optica. J. Cell. Mol. Med. 2018, 22, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Assentoft, M.; Kaptan, S.; Fenton, R.A.; Hua, S.Z.; de Groot, B.L.; MacAulay, N. Phosphorylation of rat aquaporin-4 at Ser(111) is not required for channel gating. Glia 2013, 61, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.D.; Yeh, R.; Sandstrom, A.; Chorny, I.; Harries, W.E.; Robbins, R.A.; Miercke, L.J.; Stroud, R.M. Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance. Proc. Natl. Acad. Sci. USA 2009, 106, 7437–7442. [Google Scholar] [CrossRef] [PubMed]
- Alberga, D.; Nicolotti, O.; Lattanzi, G.; Nicchia, G.P.; Frigeri, A.; Pisani, F.; Benfenati, V.; Mangiatordi, G.F. A new gating site in human aquaporin-4: Insights from molecular dynamics simulations. Biochim. Biophys. Acta 2014, 1838, 3052–3060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisani, F.; Sparaneo, A.; Tortorella, C.; Ruggieri, M.; Trojano, M.; Mola, M.G.; Nicchia, G.P.; Frigeri, A.; Svelto, M. Aquaporin-4 autoantibodies in Neuromyelitis Optica: AQP4 isoform-dependent sensitivity and specificity. PLoS ONE 2013, 8, e79185. [Google Scholar] [CrossRef]
- Kitley, J.; Woodhall, M.; Leite, M.I.; Palace, J.; Vincent, A.; Waters, P. Aquaporin-4 antibody isoform binding specificities do not explain clinical variations in NMO. Neurol. Neuroimmunol. Neuroinflamm. 2015, 2, e121. [Google Scholar] [CrossRef]
- Tuller, F.; Holzer, H.; Schanda, K.; Aboulenein-Djamshidian, F.; Hoftberger, R.; Khalil, M.; Seifert-Held, T.; Leutmezer, F.; Berger, T.; Reindl, M. Characterization of the binding pattern of human aquaporin-4 autoantibodies in patients with neuromyelitis optica spectrum disorders. J. Neuroinflamm. 2016, 13, 176. [Google Scholar] [CrossRef]
- Majed, M.; Fryer, J.P.; McKeon, A.; Lennon, V.A.; Pittock, S.J. Clinical utility of testing AQP4-IgG in CSF: Guidance for physicians. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e231. [Google Scholar] [CrossRef] [PubMed]
- Jarius, S.; Paul, F.; Fechner, K.; Ruprecht, K.; Kleiter, I.; Franciotta, D.; Ringelstein, M.; Pache, F.; Aktas, O.; Wildemann, B. Aquaporin-4 antibody testing: Direct comparison of M1-AQP4-DNA-transfected cells with leaky scanning versus M23-AQP4-DNA-transfected cells as antigenic substrate. J. Neuroinflamm. 2014, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Jarius, S.; Wildemann, B. Aquaporin-4 antibodies (NMO-IgG) as a serological marker of neuromyelitis optica: A critical review of the literature. Brain Pathol. 2013, 23, 661–683. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Tate, C.G. Quality control in eukaryotic membrane protein overproduction. J. Mol. Biol. 2014, 426, 4139–4154. [Google Scholar] [CrossRef] [PubMed]
- Wingerchuk, D.M.; Lennon, V.A.; Pittock, S.J.; Lucchinetti, C.F.; Weinshenker, B.G. Revised diagnostic criteria for neuromyelitis optica. Neurology 2006, 66, 1485–1489. [Google Scholar] [CrossRef] [Green Version]
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011, 69, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Nicchia, G.P.; Rossi, A.; Mola, M.G.; Pisani, F.; Stigliano, C.; Basco, D.; Mastrototaro, M.; Svelto, M.; Frigeri, A. Higher order structure of aquaporin-4. Neuroscience 2010, 168, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Schagger, H.; Cramer, W.A.; von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994, 217, 220–230. [Google Scholar] [CrossRef]
- Mola, M.G.; Nicchia, G.P.; Svelto, M.; Spray, D.C.; Frigeri, A. Automated cell-based assay for screening of aquaporin inhibitors. Anal. Chem. 2009, 81, 8219–8229. [Google Scholar] [CrossRef]
- Pisani, F.; Settanni, P.; Rosito, S.; Mola, M.G.; Iorio, R.; Tortorella, C.; Ruggieri, M.; Trojano, M.; Svelto, M.; Frigeri, A.; et al. Development of an Aquaporin-4 Orthogonal Array of Particle-Based ELISA for Neuromyelitis Optica Autoantibodies Detection. PLoS ONE 2015, 10, e0143679. [Google Scholar] [CrossRef]
- Pisani, F.; Rossi, A.; Nicchia, G.P.; Svelto, M.; Frigeri, A. Translational regulation mechanisms of aquaporin-4 supramolecular organization in astrocytes. Glia 2011, 59, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- Furman, C.S.; Gorelick-Feldman, D.A.; Davidson, K.G.; Yasumura, T.; Neely, J.D.; Agre, P.; Rash, J.E. Aquaporin-4 square array assembly: Opposing actions of M1 and M23 isoforms. Proc. Natl. Acad. Sci. USA 2003, 100, 13609–13614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.L.; Newstead, S. Membrane Protein Crystallisation: Current Trends and Future Perspectives. Adv. Exp. Med. Biol. 2016, 922, 61–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicchia, G.P.; Rossi, A.; Mola, M.G.; Procino, G.; Frigeri, A.; Svelto, M. Actin cytoskeleton remodeling governs aquaporin-4 localization in astrocytes. Glia 2008, 56, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Nicchia, G.P.; Frigeri, A.; Liuzzi, G.M.; Svelto, M. Inhibition of aquaporin-4 expression in astrocytes by RNAi determines alteration in cell morphology, growth, and water transport and induces changes in ischemia-related genes. FASEB J. 2003, 17, 1508–1510. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.M.; Van Hoek, A.N.; Skach, W.R.; Verkman, A.S. Aquaporin-4 dynamics in orthogonal arrays in live cells visualized by quantum dot single particle tracking. Mol. Biol. Cell 2008, 19, 3369–3378. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Goda, W.; Ikeshima-Kataoka, H.; Yasui, M. The dual effects of the astrocyte-specific enhancer of the AQP4 M1 promoter. FEBS Lett. 2017, 591, 3906–3915. [Google Scholar] [CrossRef] [PubMed]
- Neely, J.D.; Amiry-Moghaddam, M.; Ottersen, O.P.; Froehner, S.C.; Agre, P.; Adams, M.E. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc. Natl. Acad. Sci. USA 2001, 98, 14108–14113. [Google Scholar] [CrossRef] [Green Version]
- Nicchia, G.P.; Rossi, A.; Nudel, U.; Svelto, M.; Frigeri, A. Dystrophin-dependent and -independent AQP4 pools are expressed in the mouse brain. Glia 2008, 56, 869–876. [Google Scholar] [CrossRef]
- Hayakawa, S.; Mori, M.; Okuta, A.; Kamegawa, A.; Fujiyoshi, Y.; Yoshiyama, Y.; Mitsuoka, K.; Ishibashi, K.; Sasaki, S.; Hattori, T.; et al. Neuromyelitis optica and anti-aquaporin-4 antibodies measured by an enzyme-linked immunosorbent assay. J. Neuroimmunol. 2008, 196, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Jung, S.W.; Kim, Y.; Park, Y.J.; Han, K.; Oh, E.J. Detection of anti-aquaporin-4 antibodies in neuromyelitis optica: Comparison of tissue-based and cell-based indirect immunofluorescence assays and ELISA. J. Clin. Lab. Anal. 2012, 26, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Green, M.; Gilden, D.; Lam, C.; Bautista, K.; Bennett, J.L. Identification of peptide targets in neuromyelitis optica. J. Neuroimmunol. 2011, 236, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampylafka, E.I.; Routsias, J.G.; Alexopoulos, H.; Dalakas, M.C.; Moutsopoulos, H.M.; Tzioufas, A.G. Fine specificity of antibodies against AQP4: Epitope mapping reveals intracellular epitopes. J. Autoimmun. 2011, 36, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; van Hoek, A.N.; Verkman, A.S. Very high single channel water permeability of aquaporin-4 in baculovirus-infected insect cells and liposomes reconstituted with purified aquaporin-4. Biochemistry 1997, 36, 7625–7632. [Google Scholar] [CrossRef] [PubMed]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisani, F.; Simone, L.; Mola, M.G.; De Bellis, M.; Mastrapasqua, M.; Ruggieri, M.; Trojano, M.; Nicchia, G.P.; Svelto, M.; Frigeri, A. Host-Cell Type Dependent Features of Recombinant Human Aquaporin-4 Orthogonal Arrays of Particles—New Insights for Structural and Functional Studies. Cells 2019, 8, 119. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8020119
Pisani F, Simone L, Mola MG, De Bellis M, Mastrapasqua M, Ruggieri M, Trojano M, Nicchia GP, Svelto M, Frigeri A. Host-Cell Type Dependent Features of Recombinant Human Aquaporin-4 Orthogonal Arrays of Particles—New Insights for Structural and Functional Studies. Cells. 2019; 8(2):119. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8020119
Chicago/Turabian StylePisani, Francesco, Laura Simone, Maria Grazia Mola, Manuela De Bellis, Maria Mastrapasqua, Maddalena Ruggieri, Maria Trojano, Grazia Paola Nicchia, Maria Svelto, and Antonio Frigeri. 2019. "Host-Cell Type Dependent Features of Recombinant Human Aquaporin-4 Orthogonal Arrays of Particles—New Insights for Structural and Functional Studies" Cells 8, no. 2: 119. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8020119