Integrative Multi-Omics Analysis in Calcific Aortic Valve Disease Reveals a Link to the Formation of Amyloid-Like Deposits
Abstract
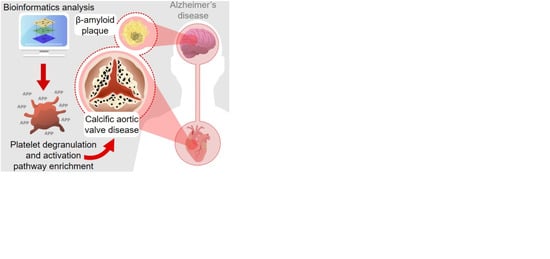
:1. Introduction
2. Materials and Methods
2.1. Study Selection and Data Collection
2.2. Pathway Over-Representation Analyses, Network Integration and Subcellular Localization
2.3. Construction of Layered Multi-Omics Network
2.4. Human Tissue
2.5. Immunohistochemistry
2.6. Thioflavin S and Congo Red Staining
2.7. Von Kossa Staining
2.8. Statistical Analysis
3. Results
3.1. Identification of Studies
3.2. Pathway Over-Representation Analysis
3.3. Complement/Coagulation Cascade and Platelet Activation/Degranulation Pathways are Over-Represented after Multi-Omics Intersection
3.4. Multi-Omics 3D Layered Network in CAVD
3.5. Calcific Aortic Valves Express Molecules Features of Amyloid Structures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yutzey, K.E.; Demer, L.L.; Body, S.C.; Huggins, G.S.; Towler, D.A.; Giachelli, C.M.; Hofmann-Bowman, M.A.; Mortlock, D.P.; Rogers, M.B.; Sadeghi, M.M.; et al. Calcific aortic valve disease: A consensus summary from the Alliance of Investigators on Calcific Aortic Valve Disease. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2387–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawade, T.A.; Newby, D.E.; Dweck, M.R. Calcification in Aortic Stenosis: The Skeleton Key. J. Am. Coll. Cardiol. 2015, 66, 561–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutcheson, J.D.; Aikawa, E.; Merryman, W.D. Potential drug targets for calcific aortic valve disease. Nat. Rev. Cardiol. 2014, 11, 218–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohler, E.R., 3rd. Are atherosclerotic processes involved in aortic-valve calcification? Lancet 2000, 356, 524–525. [Google Scholar] [CrossRef]
- Baumgartner, H.; Falk, V.; Bax, J.J.; De Bonis, M.; Hamm, C.; Holm, P.J.; Iung, B.; Lancellotti, P.; Lansac, E.; Rodriguez Munoz, D.; et al. 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur. Heart J. 2017, 38, 2739–2791. [Google Scholar] [CrossRef]
- Peeters, F.; Meex, S.J.R.; Dweck, M.R.; Aikawa, E.; Crijns, H.; Schurgers, L.J.; Kietselaer, B. Calcific aortic valve stenosis: Hard disease in the heart: A biomolecular approach towards diagnosis and treatment. Eur. Heart J. 2018, 39, 2618–2624. [Google Scholar] [CrossRef] [Green Version]
- Leon-Mimila, P.; Wang, J.; Huertas-Vazquez, A. Relevance of Multi-Omics Studies in Cardiovascular Diseases. Front. Cardiovasc. Med. 2019, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Leopold, J.A.; Maron, B.A.; Loscalzo, J. The application of big data to cardiovascular disease: Paths to precision medicine. J. Clin. Investig. 2020, 130, 29–38. [Google Scholar] [CrossRef]
- Merryman, W.D.; Schoen, F.J. Mechanisms of calcification in aortic valve disease: Role of mechanokinetics and mechanodynamics. Curr. Cardiol. Rep. 2013, 15, 355. [Google Scholar] [CrossRef]
- Trindade, F.; Ferreira, R.; Magalhaes, B.; Leite-Moreira, A.; Falcao-Pires, I.; Vitorino, R. How to use and integrate bioinformatics tools to compare proteomic data from distinct conditions? A tutorial using the pathological similarities between Aortic Valve Stenosis and Coronary Artery Disease as a case-study. J. Proteom. 2018, 171, 37–52. [Google Scholar] [CrossRef]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Orre, L.M.; Vesterlund, M.; Pan, Y.; Arslan, T.; Zhu, Y.; Fernandez Woodbridge, A.; Frings, O.; Fredlund, E.; Lehtio, J. SubCellBarCode: Proteome-wide Mapping of Protein Localization and Relocalization. Mol. Cell 2019, 73, 166–182.e167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Xia, J. Using OmicsNet for Network Integration and 3D Visualization. Curr. Protoc. Bioinform. 2019, 65, e69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlotter, F.; Halu, A.; Goto, S.; Blaser, M.C.; Body, S.C.; Lee, L.H.; Higashi, H.; DeLaughter, D.M.; Hutcheson, J.D.; Vyas, P.; et al. Spatiotemporal Multi-Omics Mapping Generates a Molecular Atlas of the Aortic Valve and Reveals Networks Driving Disease. Circulation 2018, 138, 377–393. [Google Scholar] [CrossRef] [PubMed]
- van Geemen, D.; Soares, A.L.; Oomen, P.J.; Driessen-Mol, A.; Janssen-van den Broek, M.W.; van den Bogaerdt, A.J.; Bogers, A.J.; Goumans, M.J.; Baaijens, F.P.; Bouten, C.V. Age-Dependent Changes in Geometry, Tissue Composition and Mechanical Properties of Fetal to Adult Cryopreserved Human Heart Valves. PLoS ONE 2016, 11, e0149020. [Google Scholar] [CrossRef] [PubMed]
- Oomen, P.J.A.; Loerakker, S.; van Geemen, D.; Neggers, J.; Goumans, M.T.H.; van den Bogaerdt, A.J.; Bogers, A.; Bouten, C.V.C.; Baaijens, F.P.T. Age-dependent changes of stress and strain in the human heart valve and their relation with collagen remodeling. Acta Biomater. 2016, 29, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Shi, J.; Li, B.; Zhou, Q.; Kong, X.; Bei, Y. MicroRNA Expression Signature in Human Calcific Aortic Valve Disease. Biomed. Res. Int. 2017, 2017, 4820275. [Google Scholar] [CrossRef] [Green Version]
- Coffey, S.; Williams, M.J.; Phillips, L.V.; Jones, G.T. Circulating microRNA Profiling Needs Further Refinement Before Clinical Use in Patients with Aortic Stenosis. J. Am. Heart Assoc. 2015, 4, e002150. [Google Scholar] [CrossRef] [Green Version]
- Ohukainen, P.; Syvaranta, S.; Napankangas, J.; Rajamaki, K.; Taskinen, P.; Peltonen, T.; Helske-Suihko, S.; Kovanen, P.T.; Ruskoaho, H.; Rysa, J. MicroRNA-125b and chemokine CCL4 expression are associated with calcific aortic valve disease. Ann. Med. 2015, 47, 423–429. [Google Scholar] [CrossRef]
- Shi, J.; Liu, H.; Wang, H.; Kong, X. MicroRNA Expression Signature in Degenerative Aortic Stenosis. Biomed. Res. Int. 2016, 2016, 4682172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffey, S.; Williams, M.J.; Phillips, L.V.; Galvin, I.F.; Bunton, R.W.; Jones, G.T. Integrated microRNA and messenger RNA analysis in aortic stenosis. Sci. Rep. 2016, 6, 36904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anger, T.; Carson, W.; Weyand, M.; Daniel, W.G.; Hoeher, M.; Garlichs, C.D. Atherosclerotic inflammation triggers osteogenic bone transformation in calcified and stenotic human aortic valves: Still a matter of debate. Exp. Mol. Pathol. 2009, 86, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Bosse, Y.; Miqdad, A.; Fournier, D.; Pepin, A.; Pibarot, P.; Mathieu, P. Refining molecular pathways leading to calcific aortic valve stenosis by studying gene expression profile of normal and calcified stenotic human aortic valves. Circ. Cardiovasc. Genet. 2009, 2, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Guauque-Olarte, S.; Droit, A.; Tremblay-Marchand, J.; Gaudreault, N.; Kalavrouziotis, D.; Dagenais, F.; Seidman, J.G.; Body, S.C.; Pibarot, P.; Mathieu, P.; et al. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol. Genom. 2016, 48, 749–761. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Xuan, C.; Yang, Q.; Liu, X.C.; Liu, Z.G.; He, G.W. Identification of altered plasma proteins by proteomic study in valvular heart diseases and the potential clinical significance. PLoS ONE 2013, 8, e72111. [Google Scholar] [CrossRef] [Green Version]
- Gil-Dones, F.; Darde, V.M.; Alonso-Orgaz, S.; Lopez-Almodovar, L.F.; Mourino-Alvarez, L.; Padial, L.R.; Vivanco, F.; Barderas, M.G. Inside human aortic stenosis: A proteomic analysis of plasma. J. Proteom. 2012, 75, 1639–1653. [Google Scholar] [CrossRef]
- Martin-Rojas, T.; Gil-Dones, F.; Lopez-Almodovar, L.F.; Padial, L.R.; Vivanco, F.; Barderas, M.G. Proteomic profile of human aortic stenosis: Insights into the degenerative process. J. Proteome Res. 2012, 11, 1537–1550. [Google Scholar] [CrossRef]
- Martin-Rojas, T.; Mourino-Alvarez, L.; Alonso-Orgaz, S.; Rosello-Lleti, E.; Calvo, E.; Lopez-Almodovar, L.F.; Rivera, M.; Padial, L.R.; Lopez, J.A.; de la Cuesta, F.; et al. iTRAQ proteomic analysis of extracellular matrix remodeling in aortic valve disease. Sci. Rep. 2015, 5, 17290. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, K.; Satoh, K.; Maniwa, T.; Araki, A.; Maruyama, R.; Oda, T. Noticeable decreased expression of tenascin-X in calcific aortic valves. Connect. Tissue Res. 2012, 53, 460–468. [Google Scholar] [CrossRef]
- Mourino-Alvarez, L.; Baldan-Martin, M.; Gonzalez-Calero, L.; Martinez-Laborde, C.; Sastre-Oliva, T.; Moreno-Luna, R.; Lopez-Almodovar, L.F.; Sanchez, P.L.; Fernandez-Aviles, F.; Vivanco, F.; et al. Patients with calcific aortic stenosis exhibit systemic molecular evidence of ischemia, enhanced coagulation, oxidative stress and impaired cholesterol transport. Int. J. Cardiol. 2016, 225, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Olkowicz, M.; Debski, J.; Jablonska, P.; Dadlez, M.; Smolenski, R.T. Application of a new procedure for liquid chromatography/mass spectrometry profiling of plasma amino acid-related metabolites and untargeted shotgun proteomics to identify mechanisms and biomarkers of calcific aortic stenosis. J. Chromatogr. A 2017, 1517, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Rehulkova, H.; Rehulka, P.; Myslivcova Fucikova, A.; Stulik, J.; Pudil, R. Identification of novel biomarker candidates for hypertrophic cardiomyopathy and other cardiovascular diseases leading to heart failure. Physiol. Res. 2016, 65, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Chikada, M.; Yokoyama, M.K.; Kurokawa, M.S.; Ando, T.; Furukawa, H.; Arito, M.; Miyairi, T.; Kato, T. Aberrant Glycosylation of Lumican in Aortic Valve Stenosis Revealed by Proteomic Analysis. Int. Heart J. 2016, 57, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Howie, A.J. Origins of a pervasive, erroneous idea: The “green birefringence” of Congo red-stained amyloid. Int. J. Exp. Pathol. 2019, 100, 208–221. [Google Scholar] [CrossRef]
- Klatskin, G. Nonspecific Green Birefringence in Congo Red-Stained Tissues. Am. J. Pathol. 1969, 56, 1. [Google Scholar]
- Breyne, J.; Juthier, F.; Corseaux, D.; Marechaux, S.; Zawadzki, C.; Jeanpierre, E.; Ung, A.; Ennezat, P.V.; Susen, S.; Van Belle, E.; et al. Atherosclerotic-like process in aortic stenosis: Activation of the tissue factor-thrombin pathway and potential role through osteopontin alteration. Atherosclerosis 2010, 213, 369–376. [Google Scholar] [CrossRef]
- Helske, S.; Oksjoki, R.; Lindstedt, K.A.; Lommi, J.; Turto, H.; Werkkala, K.; Kupari, M.; Kovanen, P.T. Complement system is activated in stenotic aortic valves. Atherosclerosis 2008, 196, 190–200. [Google Scholar] [CrossRef]
- Kapusta, P.; Wypasek, E.; Natorska, J.; Grudzien, G.; Sobczyk, D.; Sadowski, J.; Undas, A. Factor XIII expression within aortic valves and its plasma activity in patients with aortic stenosis: Association with severity of disease. Thromb. Haemost. 2012, 108, 1172–1179. [Google Scholar] [CrossRef]
- Natorska, J.; Marek, G.; Hlawaty, M.; Sadowski, J.; Tracz, W.; Undas, A. Fibrin presence within aortic valves in patients with aortic stenosis: Association with in vivo thrombin generation and fibrin clot properties. Thromb. Haemost. 2011, 105, 254–260. [Google Scholar] [CrossRef]
- Borissoff, J.I.; Heeneman, S.; Kilinc, E.; Kassak, P.; Van Oerle, R.; Winckers, K.; Govers-Riemslag, J.W.; Hamulyak, K.; Hackeng, T.M.; Daemen, M.J.; et al. Early atherosclerosis exhibits an enhanced procoagulant state. Circulation 2010, 122, 821–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nording, H.; Langer, H.F. Complement links platelets to innate immunity. Semin. Immunol. 2018, 37, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Conway, E.M. Platelets and Complement Cross-Talk in Early Atherogenesis. Front. Cardiovasc. Med. 2019, 6, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincentelli, A.; Susen, S.; Le Tourneau, T.; Six, I.; Fabre, O.; Juthier, F.; Bauters, A.; Decoene, C.; Goudemand, J.; Prat, A.; et al. Acquired von Willebrand syndrome in aortic stenosis. N. Engl. J. Med. 2003, 349, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Bouchareb, R.; Boulanger, M.C.; Tastet, L.; Mkannez, G.; Nsaibia, M.J.; Hadji, F.; Dahou, A.; Messadeq, Y.; Arsenault, B.J.; Pibarot, P.; et al. Activated platelets promote an osteogenic programme and the progression of calcific aortic valve stenosis. Eur. Heart J. 2019, 40, 1362–1373. [Google Scholar] [CrossRef]
- Zucoloto, A.Z.; Jenne, C.N. Platelet-Neutrophil Interplay: Insights into Neutrophil Extracellular Trap (NET)-Driven Coagulation in Infection. Front. Cardiovasc. Med. 2019, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Coughlin, S.R. How the protease thrombin talks to cells. Proc. Natl. Acad. Sci. USA 1999, 96, 11023–11027. [Google Scholar] [CrossRef] [Green Version]
- Rouzaud-Laborde, C.; Delmas, C.; Pizzinat, N.; Tortosa, F.; Garcia, C.; Mialet-Perez, J.; Payrastre, B.; Sie, P.; Spreux-Varoquaux, O.; Sallerin, B.; et al. Platelet activation and arterial peripheral serotonin turnover in cardiac remodeling associated to aortic stenosis. Am. J. Hematol. 2015, 90, 15–19. [Google Scholar] [CrossRef]
- Hutcheson, J.D.; Ryzhova, L.M.; Setola, V.; Merryman, W.D. 5-HT(2B) antagonism arrests non-canonical TGF-beta1-induced valvular myofibroblast differentiation. J. Mol. Cell Cardiol. 2012, 53, 707–714. [Google Scholar] [CrossRef] [Green Version]
- Rothman, R.B.; Baumann, M.H. Serotonergic drugs and valvular heart disease. Expert Opin. Drug Saf. 2009, 8, 317–329. [Google Scholar] [CrossRef]
- Liu, A.C.; Joag, V.R.; Gotlieb, A.I. The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. Am. J. Pathol. 2007, 171, 1407–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helske, S.; Syvaranta, S.; Lindstedt, K.A.; Lappalainen, J.; Oorni, K.; Mayranpaa, M.I.; Lommi, J.; Turto, H.; Werkkala, K.; Kupari, M.; et al. Increased expression of elastolytic cathepsins S, K, and V and their inhibitor cystatin C in stenotic aortic valves. Arter. Thromb. Vasc. Biol. 2006, 26, 1791–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kossar, A.P.; Anselmo, W.; Grau, J.B.; Liu, Y.; Small, A.; Carter, S.L.; Salvador, L.; Zhao, L.; Cvijic, M.E.; Li, Z.; et al. Circulating and tissue matricellular RNA and protein expression in calcific aortic valve disease. Physiol. Genom. 2020, 52, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Tublin, J.M.; Adelstein, J.M.; Del Monte, F.; Combs, C.K.; Wold, L.E. Getting to the Heart of Alzheimer Disease. Circ. Res. 2019, 124, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Troncone, L.; Luciani, M.; Coggins, M.; Wilker, E.H.; Ho, C.Y.; Codispoti, K.E.; Frosch, M.P.; Kayed, R.; Del Monte, F. Abeta Amyloid Pathology Affects the Hearts of Patients with Alzheimer’s Disease: Mind the Heart. J. Am. Coll. Cardiol. 2016, 68, 2395–2407. [Google Scholar] [CrossRef] [PubMed]
- Reitz, C.; Brickman, A.M.; Luchsinger, J.A.; Wu, W.E.; Small, S.A.; Tang, M.; Mayeux, R. Frequency of Subclinical Heart Disease in Elderly Persons with Dementia. Am. J. Geriatr. Cardiol. 2007, 16, 183–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novaro, G.M.; Sachar, R.; Pearce, G.L.; Sprecher, D.L.; Griffin, B.P. Association between apolipoprotein E alleles and calcific valvular heart disease. Circulation 2003, 108, 1804–1808. [Google Scholar] [CrossRef]
- Wolters, F.J.; Bos, D.; Vernooij, M.W.; Franco, O.H.; Hofman, A.; Koudstaal, P.J.; van der Lugt, A.; Ikram, M.A. Aortic Valve Calcification and the Risk of dementia: A Population-Based Study. J. Alzheimer’s Dis. 2016, 55, 893–897. [Google Scholar] [CrossRef]
- van der Kant, R.; Goldstein, L.S. Cellular functions of the amyloid precursor protein from development to dementia. Dev. Cell 2015, 32, 502–515. [Google Scholar] [CrossRef] [Green Version]
- Paula-Lima, A.C.; Tricerri, M.A.; Brito-Moreira, J.; Bomfim, T.R.; Oliveira, F.F.; Magdesian, M.H.; Grinberg, L.T.; Panizzutti, R.; Ferreira, S.T. Human apolipoprotein A-I binds amyloid-beta and prevents Abeta-induced neurotoxicity. Int. J. Biochem. Cell Biol. 2009, 41, 1361–1370. [Google Scholar] [CrossRef]
- Raditsis, A.V.; Milojevic, J.; Melacini, G. Abeta association inhibition by transferrin. Biophys. J. 2013, 105, 473–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyatt, A.R.; Constantinescu, P.; Ecroyd, H.; Dobson, C.M.; Wilson, M.R.; Kumita, J.R.; Yerbury, J.J. Protease-activated alpha-2-macroglobulin can inhibit amyloid formation via two distinct mechanisms. FEBS Lett. 2013, 587, 398–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.S.; Eira, J.; Ribeiro, C.A.; Oliveira, A.; Sousa, M.M.; Cardoso, I.; Liz, M.A. Transthyretin neuroprotection in Alzheimer’s disease is dependent on proteolysis. Neurobiol. Aging 2017, 59, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, J.N.; Ye, Z.; Reixach, N.; Friske, L.; Levy, C.; Das, P.; Golde, T.; Masliah, E.; Roberts, A.R.; Bartfai, T. Transthyretin protects Alzheimer’s mice from the behavioral and biochemical effects of Abeta toxicity. Proc. Natl. Acad. Sci. USA 2008, 105, 2681–2686. [Google Scholar] [CrossRef] [Green Version]
- Stein, T.D.; Anders, N.J.; DeCarli, C.; Chan, S.L.; Mattson, M.P.; Johnson, J.A. Neutralization of transthyretin reverses the neuroprotective effects of secreted amyloid precursor protein (APP) in APPSW mice resulting in tau phosphorylation and loss of hippocampal neurons: Support for the amyloid hypothesis. J. Neurosci. 2004, 24, 7707–7717. [Google Scholar] [CrossRef] [Green Version]
- Pedrero-Prieto, C.M.; Flores-Cuadrado, A.; Saiz-Sanchez, D.; Ubeda-Banon, I.; Frontinan-Rubio, J.; Alcain, F.J.; Mateos-Hernandez, L.; de la Fuente, J.; Duran-Prado, M.; Villar, M.; et al. Human amyloid-beta enriched extracts: Evaluation of in vitro and in vivo internalization and molecular characterization. Alzheimers Res. Ther. 2019, 11, 56. [Google Scholar] [CrossRef]
- Zamolodchikov, D.; Renne, T.; Strickland, S. The Alzheimer’s disease peptide beta-amyloid promotes thrombin generation through activation of coagulation factor XII. J. Thromb. Haemost. 2016, 14, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- Hur, W.S.; Mazinani, N.; Lu, X.J.D.; Yefet, L.S.; Byrnes, J.R.; Ho, L.; Yeon, J.H.; Filipenko, S.; Wolberg, A.S.; Jefferies, W.A.; et al. Coagulation factor XIIIa cross-links amyloid β into dimers and oligomers and to blood proteins. J. Biol. Chem. 2019, 294, 390–396. [Google Scholar] [CrossRef] [Green Version]
- Strickland, S. Blood will out: Vascular contributions to Alzheimer’s disease. J. Clin. Investig. 2018, 128, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Cortes-Canteli, M.; Iadecola, C. Alzheimer’s Disease and Vascular Aging: JACC Focus Seminar. J. Am. Coll. Cardiol. 2020, 75, 942–951. [Google Scholar] [CrossRef]
- Evin, G.; Li, Q.X. Platelets and Alzheimer’s disease: Potential of APP as a biomarker. World J. Psychiatry 2012, 2, 102–113. [Google Scholar] [CrossRef]
- De Meyer, G.R.; De Cleen, D.M.; Cooper, S.; Knaapen, M.W.; Jans, D.M.; Martinet, W.; Herman, A.G.; Bult, H.; Kockx, M.M. Platelet phagocytosis and processing of beta-amyloid precursor protein as a mechanism of macrophage activation in atherosclerosis. Circ. Res. 2002, 90, 1197–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Parre, T.J.; Guns, P.J.; Fransen, P.; Martinet, W.; Bult, H.; Herman, A.G.; De Meyer, G.R. Attenuated atherogenesis in apolipoprotein E-deficient mice lacking amyloid precursor protein. Atherosclerosis 2011, 216, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Tibolla, G.; Norata, G.D.; Meda, C.; Arnaboldi, L.; Uboldi, P.; Piazza, F.; Ferrarese, C.; Corsini, A.; Maggi, A.; Vegeto, E.; et al. Increased atherosclerosis and vascular inflammation in APP transgenic mice with apolipoprotein E deficiency. Atherosclerosis 2010, 210, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Kong, G.K.; Adams, J.J.; Harris, H.H.; Boas, J.F.; Curtain, C.C.; Galatis, D.; Masters, C.L.; Barnham, K.J.; McKinstry, W.J.; Cappai, R.; et al. Structural studies of the Alzheimer’s amyloid precursor protein copper-binding domain reveal how it binds copper ions. J. Mol. Biol. 2007, 367, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.H. Localized Dystrophic Amyloidosis of Heart Valves. Hum. Pathol. 1983, 14, 649–653. [Google Scholar] [CrossRef]
- Falk, E.; Ladefoged, C.; Rohr, C. Amyloid deposits in aortic and mitral valves. Virchows Arch. Pathol. Anat. 1981, 404, 301–312. [Google Scholar]
- Goffin, Y. Microscopic amyloid deposits in the heart valves: A common local complication of chronic damage and scarring. J. Clin. Pathol. 1980, 33, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Audet, A.; Cote, N.; Couture, C.; Bosse, Y.; Despres, J.P.; Pibarot, P.; Mathieu, P. Amyloid substance within stenotic aortic valves promotes mineralization. Histopathology 2012, 61, 610–619. [Google Scholar] [CrossRef]
- Clement, C.G.; Truong, L.D. An evaluation of Congo red fluorescence for the diagnosis of amyloidosis. Hum. Pathol. 2014, 45, 1766–1772. [Google Scholar] [CrossRef]
- Nitsche, C.; Aschauer, S.; Kammerlander, A.A.; Schneider, M.; Poschner, T.; Duca, F.; Binder, C.; Koschutnik, M.; Stiftinger, J.; Goliasch, G.; et al. Light-chain and transthyretin cardiac amyloidosis in severe aortic stenosis: Prevalence, screening possibilities, and outcome. Eur. J. Heart Fail. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesko, G.; Jenei, Z.; Varga, G.; Apor, A.; Vago, H.; Czibor, S.; Prohaszka, Z.; Masszi, T.; Pozsonyi, Z. Coexistence of aortic valve stenosis and cardiac amyloidosis: Echocardiographic and clinical significance. Cardiovasc. Ultrasound 2019, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ternacle, J.; Krapf, L.; Mohty, D.; Magne, J.; Nguyen, A.; Galat, A.; Gallet, R.; Teiger, E.; Cote, N.; Clavel, M.A.; et al. Aortic Stenosis and Cardiac Amyloidosis. J. Am. Coll. Cardiol. 2019, 74, 2638–2651. [Google Scholar] [CrossRef] [PubMed]
- Emdin, M.; Aimo, A.; Rapezzi, C.; Fontana, M.; Perfetto, F.; Seferovic, P.M.; Barison, A.; Castiglione, V.; Vergaro, G.; Giannoni, A.; et al. Treatment of cardiac transthyretin amyloidosis: An update. Eur. Heart J. 2019, 40, 3699–3706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, K.D. Pathogenesis of calcific aortic valve disease: A disease process comes of age (and a good deal more). Arter. Thromb. Vasc. Biol. 2006, 26, 1721–1728. [Google Scholar] [CrossRef] [Green Version]
- Chawade, A.; Alexandersson, E.; Levander, F. Normalyzer: A tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 2014, 13, 3114–3120. [Google Scholar] [CrossRef]
- Salvador-Olivan, J.A.; Marco-Cuenca, G.; Arquero-Aviles, R. Errors in search strategies used in systematic reviews and their effects on information retrieval. J. Med. Libr. Assoc. 2019, 107, 210–221. [Google Scholar] [CrossRef] [Green Version]
- Sonawane, A.R.; Weiss, S.T.; Glass, K.; Sharma, A. Network Medicine in the Age of Biomedical Big Data. Front. Genet. 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]






| Total | miRNA | mRNA | Protein | |||||
|---|---|---|---|---|---|---|---|---|
| Studies, n | 20 | 5 | 5 | 10 | ||||
| Control | CAVD | Control | CAVD | Control | CAVD | Control | CAVD | |
| Samples, n | 270 | 258 | 56 | 52 | 29 | 34 | 185 | 172 |
| Samples used for omics, n | 197 | 172 | 56 | 50 | 29 | 32 | 112 | 90 |
| Age 1, mean ± SD | 59.0 ± 10.8 | 68.3 ± 8.2 | 51.9 ± 11.7 | 67.8 ± 11.8 | 50.9 ± 6.1 | 63.3 ± 5.1 | 65.7 ± 7.5 | 70.8 ± 7.2 |
| p-value | p = 0.008 | p = 0.105 | p = 0.021 | p = 0.166 | ||||
| Male 1, % ± SD | 69.8 ± 25.6 | 67.3 ± 26.1 | 81.3 ± 23.9 | 80.8 ± 22.3 | 91.7 ± 16.7 | 85.0 ± 30.0 | 53.2 ± 19.4 | 53.4 ±19.3 |
| Source of calcified AV, studies/overall studies | ||||||||
| AV replacement surgery | 14/20 | 4/5 | 5/5 | 5/10 | ||||
| Source of control AV, studies/overall studies | ||||||||
| Aortic regurgitation | 3/20 | 1/5 | 2/5 | |||||
| Autopsy | 5/20 | 1/5 | 1/5 | 3/10 | ||||
| Transplantation | 4/20 | 2/5 | 2/5 | |||||
| Non-calcified tissue part | 2/20 | 2/10 | ||||||
| Source of plasma, studies/overall studies | ||||||||
| Patients with AS | 6/20 | 1/5 | 5/10 | |||||
| Subjects without CVD | 5/20 | 1/5 | 4/10 | |||||
| Aortic regurgitation, non-AS | 1/20 | 1/10 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heuschkel, M.A.; Skenteris, N.T.; Hutcheson, J.D.; van der Valk, D.D.; Bremer, J.; Goody, P.; Hjortnaes, J.; Jansen, F.; Bouten, C.V.C.; van den Bogaerdt, A.; et al. Integrative Multi-Omics Analysis in Calcific Aortic Valve Disease Reveals a Link to the Formation of Amyloid-Like Deposits. Cells 2020, 9, 2164. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9102164
Heuschkel MA, Skenteris NT, Hutcheson JD, van der Valk DD, Bremer J, Goody P, Hjortnaes J, Jansen F, Bouten CVC, van den Bogaerdt A, et al. Integrative Multi-Omics Analysis in Calcific Aortic Valve Disease Reveals a Link to the Formation of Amyloid-Like Deposits. Cells. 2020; 9(10):2164. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9102164
Chicago/Turabian StyleHeuschkel, Marina A., Nikolaos T. Skenteris, Joshua D. Hutcheson, Dewy D. van der Valk, Juliane Bremer, Philip Goody, Jesper Hjortnaes, Felix Jansen, Carlijn V.C. Bouten, Antoon van den Bogaerdt, and et al. 2020. "Integrative Multi-Omics Analysis in Calcific Aortic Valve Disease Reveals a Link to the Formation of Amyloid-Like Deposits" Cells 9, no. 10: 2164. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9102164