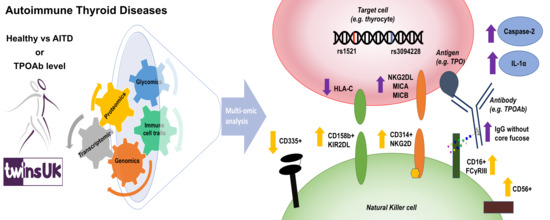
Dysregulated Antibody, Natural Killer Cell and Immune Mediator Profiles in Autoimmune Thyroid Diseases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sample
2.2. Data Statement
2.3. Definition of AITD and Detection of TSH and TPOAb
2.4. Detection of IgG Glycosylation Profiling for Discovery
2.5. Detection of Immune Cell Traits
2.6. Detection of Protein Profiling in Plasma
2.7. Statistical Analyses
2.8. Genome-Wide Association Analysis on IgG N-Glycan Traits
2.9. Determination of Shared Genetic Variants and Genes between IgG N-Glycan Traits, Immune Cell Traits, Protein Abundance, and Thyroid Functions and Diseases
2.10. Visualization
3. Results
3.1. Depletion of IgG Core Fucose is Positively Associated with Increased CD158b+ CD314+ CD335− NK Cell Subset Counts
3.2. The AITD-Associated Genetic Variants, rs1521 and rs3094228, Alter Thyroid Cell Expression of Ligands for CD314 and CD158b Immunoreceptors
3.3. AITD is Associated with Increased Serum Caspase-2 and IL-1α
3.4. Afucosylated IgG is Associated with Serum Levels of Several Circulating Proteins
3.5. Free-Soluble Plasma Desmoglein-2 Protein is Associated with AITD Genetic Variants and Two AITD-IgG N-Glycan Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
Appendix A
Appendix A.1. Detection of TSH and TPOAb in Human Sera
Appendix A.2. Detection of IgG Glycosylation Profiling
Appendix A.3. Detection of Protein Profiling in Plasma
Appendix A.4. Selection of SNPs Associated with Immune Cell Traits
Appendix A.5. Determination of Effective Number of Independent Tests for Different -omic Data
Appendix A.6. Association Studies between -omics Features and Thyroid Phenotypes
Appendix A.7. Heritability Analysis for Proteins
References
- Brix, T.H.; Kyvik, K.O.; Christensen, K.; Hegedüs, L. Evidence for a Major Role of Heredity in Graves’ Disease: A Population-Based Study of Two Danish Twin Cohorts. J. Clin. Endocrinol. Metab. 2001, 86, 930–934. [Google Scholar] [CrossRef]
- Hansen, P.S.; Brix, T.H.; Bennedbæk, F.N.; Bonnema, S.J.; Iachine, I.; Kyvik, K.O.; Hegedüs, L. The relative importance of genetic and environmental factors in the aetiology of thyroid nodularity: A study of healthy Danish twins. Clin. Endocrinol. 2006, 62, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Brix, T.H.; Kyvik, K.O.; Hegedüs, L. A population-based study of chronic autoimmune hypothyroidism in Danish twins. J. Clin. Endocrinol. Metab. 2000, 85, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.C.; Šimurina, M.; Ząbczyńska, M.; Martinic Kavur, M.; Rydlewska, M.; Pezer, M.; Kozłowska, K.; Burri, A.; Vilaj, M.; Turek-Jabrocka, R.; et al. Decreased immunoglobulin G core fucosylation, a player in antibody-dependent cell-mediated cytotoxicity, is associated with autoimmune thyroid diseases. Mol. Cell. Proteom. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmonds, M.J.; Gough, S.C.L. Genetic insights into disease mechanisms of autoimmunity. Br. Med. Bull. 2005, 71, 93–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fröhlich, E.; Wahl, R. Thyroid autoimmunity: Role of anti-thyroid antibodies in thyroid and extra-thyroidal diseases. Front. Immunol. 2017, 8, 521. [Google Scholar] [CrossRef]
- Wang, B.; Shao, X.; Song, R.; Xu, D.; Zhang, J.A. The emerging role of epigenetics in autoimmune thyroid diseases. Front. Immunol. 2017, 8, 396. [Google Scholar] [CrossRef] [Green Version]
- Armengol, M.P.; Juan, M.; Lucas-Martín, A.; Fernández-Figueras, M.T.; Jaraquemada, D.; Gallart, T.; Pujol-Borrell, R. Thyroid autoimmune disease: Demonstration of thyroid antigen-specific B cells and recombination-activating gene expression in chemokine-containing active intrathyroidal germinal centers. Am. J. Pathol. 2001, 159, 861–873. [Google Scholar] [CrossRef]
- Braverman, L.; Wartofsky, L. Thyroid Tests; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2014. [Google Scholar]
- British Thyroid Association. Adapted Summary of UK Guidelines for the Use of Thyroid Function Tests; British Thyroid Association: Harrogate, UK, 2006. [Google Scholar]
- Calder, E.A.; Penhale, W.J.; McLeman, D.; Barnes, E.W.; Irvine, W.J. Lymphocyte-dependent antibody-mediated cytotoxicity in Hashimoto thyroiditis. Clin. Exp. Immunol. 1973, 14, 153–158. [Google Scholar]
- Nada, A.M.; Hammouda, M. Immunoregulatory T cells, LFA-3 and HLA-DR in autoimmune thyroid diseases. Indian J. Endocrinol. Metab. 2014, 18, 574–581. [Google Scholar] [CrossRef]
- Mikos, H.; Mikos, M.; Rabska-Pietrzak, B.; Niedziela, M. The clinical role of serum concentrations of selected cytokines: IL-1beta, TNF-alpha and IL-6 in diagnosis of autoimmune thyroid disease (AITD) in children. Autoimmunity 2014, 47, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Rodien, P.; Madec, A.M.; Ruf, J.; Rajas, F.; Bornet, H.; Carayon, P.; Orgiazzi, J. Antibody-dependent cell-mediated cytotoxicity in autoimmune thyroid disease: Relationship to antithyroperoxidase antibodies. J. Clin. Endocrinol. Metab. 1996, 81, 2595–2600. [Google Scholar] [PubMed] [Green Version]
- Metcalfe, R.A.; Oh, Y.S.; Stroud, C.; Arnold, K.; Weetman, A.P. Analysis of antibody-dependent cell-mediated cytotoxicity in autoimmune thyroid disease. Autoimmunity 1997, 25, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Maverakis, E.; Kim, K.; Shimoda, M.; Gershwin, M.E.; Patel, F.; Wilken, R.; Raychaudhuri, S.; Ruhaak, L.R.; Lebrilla, C.B. Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: A critical review. J. Autoimmun. 2015, 57, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marth, J.D.; Grewal, P.K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 2008, 8, 874–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanda, Y.; Yamada, T.; Mori, K.; Okazaki, A.; Inoue, M.; Kitajima-Miyama, K.; Kuni-Kamochi, R.; Nakano, R.; Yano, K.; Kakita, S.; et al. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: The high-mannose, hybrid, and complex types. Glycobiology 2007, 17, 104–118. [Google Scholar] [CrossRef]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Gloria Meng, Y.; Weikert, S.H.A.; Presta, L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef] [Green Version]
- Niwa, R.; Hatanaka, S.; Shoji-Hosaka, E.; Sakurada, M.; Kobayashi, Y.; Uehara, A.; Yokoi, H.; Nakamura, K.; Shitara, K. Enhancement of the antibody-dependent cellular cytotoxicity of low-fucose IgG1 Is independent of FcgammaRIIIa functional polymorphism. Clin. Cancer Res. 2004, 10, 6248–6255. [Google Scholar] [CrossRef] [Green Version]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. [Google Scholar] [CrossRef] [Green Version]
- Ferrara, C.; Grau, S.; Jäger, C.; Sondermann, P.; Brünker, P.; Waldhauer, I.; Hennig, M.; Ruf, A.; Rufer, A.C.; Stihle, M.; et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. USA 2011, 108, 12669–12674. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.T.; Ravetch, J.V. Functional diversification of IgGs through Fc glycosylation. J. Clin. Investig. 2019, 129, 3492–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niwa, R.; Sakurada, M.; Kobayashi, Y.; Uehara, A.; Matsushima, K.; Ueda, R.; Nakamura, K.; Shitara, K. Enhanced natural killer cell binding and activation by low-fucose IgG1 antibody results in potent antibody-dependent cellular cytotoxicity induction at lower antigen density. Clin. Cancer Res. 2005, 11, 2327–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ząbczyńska, M.; Link-Lenczowski, P.; Novokmet, M.; Martin, T.; Turek-Jabrocka, R.; Trofimiuk-Müldner, M.; Pocheć, E. Altered N-glycan profile of IgG-depleted serum proteins in Hashimoto’s thyroiditis. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129464. [Google Scholar] [CrossRef] [PubMed]
- Klarić, L.; Tsepilov, Y.A.; Stanton, C.M.; Mangino, M.; Sikka, T.T.; Esko, T.; Pakhomov, E.; Salo, P.; Deelen, J.; McGurnaghan, S.J.; et al. Glycosylation of immunoglobulin G is regulated by a large network of genes pleiotropic with inflammatory diseases. Sci. Adv. 2020, 6, eaax0301. [Google Scholar] [CrossRef] [Green Version]
- Mangino, M.; Beddall, M.H.; Spector, T.D.; Roederer, M.; Nestle, F.O. Innate and adaptive immune traits are differentially affected by genetic and environmental factors. Nat. Commun. 2017, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Roederer, M.; Quaye, L.; Mangino, M.; Beddall, M.H.; Mahnke, Y.; Chattopadhyay, P.; Tosi, I.; Napolitano, L.; Terranova Barberio, M.; Menni, C.; et al. The genetic architecture of the human immune system: A bioresource for autoimmunity and disease pathogenesis. Cell 2015, 161, 387–403. [Google Scholar] [CrossRef] [Green Version]
- Lauc, G.; Huffman, J.E.; Pučić, M.; Zgaga, L.; Adamczyk, B.; Mužinić, A.; Novokmet, M.; Polašek, O.; Gornik, O.; Krištić, J.; et al. Loci Associated with N-Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers. PLoS Genet. 2013, 9, e1003225. [Google Scholar] [CrossRef]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014, 42, 1001–1006. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef]
- Ardlie, K.G.; DeLuca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; Lek, M.; et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar]
- Aguet, F.; Brown, A.A.; Castel, S.E.; Davis, J.R.; He, Y.; Jo, B.; Mohammadi, P.; Park, Y.S.; Parsana, P.; Segrè, A.V.; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar]
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Brown, A.; Castel, S.E.; Jo, B.; Kasela, S.; Kim-Hellmuth, S.; Liang, Y.; Oliva, M.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv 2019. [Google Scholar] [CrossRef]
- Westra, H.J.; Arends, D.; Esko, T.; Peters, M.J.; Schurmann, C.; Schramm, K.; Kettunen, J.; Yaghootkar, H.; Fairfax, B.P.; Andiappan, A.K.; et al. Cell Specific eQTL Analysis without Sorting Cells. PLoS Genet. 2015, 11, e1005223. [Google Scholar] [CrossRef] [PubMed]
- Spector, T.D.; Williams, F.M.K. The UK Adult Twin Registry (TwinsUK). Twin Res. Hum. Genet. 2006, 9, 899–906. [Google Scholar] [CrossRef]
- Moayyeri, A.; Hammond, C.J.; Hart, D.J.; Spector, T.D. The UK Adult Twin Registry (TwinsUK Resource). Twin Res. Hum. Genet. 2013, 16, 144–149. [Google Scholar] [CrossRef] [Green Version]
- Hollowell, J.G.; Staehling, N.W.; Flanders, W.D.; Hannon, W.H.; Gunter, E.W.; Spencer, C.A.; Braverman, L.E. Serum TSH, T 4, and Thyroid Antibodies in the United States Population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J. Clin. Endocrinol. Metab. 2002, 87, 489–499. [Google Scholar] [CrossRef]
- Pearce, E.N.; Farwell, A.; Braverman, L.E. Thyroiditis. N. Engl. J. Med. 2003, 348, 2646–2655. [Google Scholar] [CrossRef]
- Elhomsy, G. Antithyroid Antibody. Available online: https://emedicine.medscape.com/article/2086819-overview (accessed on 8 January 2020).
- Menni, C.; Kiddle, S.J.; Mangino, M.; Viñuela, A.; Psatha, M.; Steves, C.; Sattlecker, M.; Buil, A.; Newhouse, S.; Nelson, S.; et al. Circulating proteomic signatures of chronological age. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 70, 809–816. [Google Scholar] [CrossRef]
- Kiddle, S.J.; Steves, C.J.; Mehta, M.; Simmons, A.; Xu, X.; Newhouse, S.; Sattlecker, M.; Ashton, N.J.; Bazenet, C.; Killick, R.; et al. Plasma protein biomarkers of Alzheimer’s disease endophenotypes in asymptomatic older twins: Early cognitive decline and regional brain volumes. Transl. Psychiatry 2015, 5, e584. [Google Scholar] [CrossRef] [Green Version]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Exffects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Karssen, L.; van Dujin, C.M.; Aulchenko, Y.S. The GenABEL Project for statistical genomics. F1000Research 2016, 5, 914. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Klarić, L.; Sharapov, S.; Mangino, M.; Ning, Z.; Wu, D.; Trbojević-Akmačić, I.; Pučić-Baković, M.; Rudan, I.; Polašek, O.; et al. Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation. Nat. Commun. 2017, 8, 447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farh, K.K.-H.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.H.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2015, 44, D877–D881. [Google Scholar] [CrossRef]
- Jennewein, M.F.; Alter, G. The Immunoregulatory Roles of Antibody Glycosylation. Trends Immunol. 2017, 38, 358–372. [Google Scholar] [CrossRef]
- Kobata, A. The N-linked sugar chains of human immunoglobulin G: Their unique pattern, and their functional roles. Biochim. Biophys. Acta 2008, 1780, 472–478. [Google Scholar] [CrossRef]
- Arnold, J.N.; Wormald, M.R.; Sim, R.B.; Rudd, P.M.; Dwek, R.A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 2007, 25, 21–50. [Google Scholar] [CrossRef]
- Subedi, G.P.; Barb, A.W. The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure 2015, 23, 1573–1583. [Google Scholar] [CrossRef] [Green Version]
- Subedi, G.P.; Barb, A.W. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. mAbs 2016, 8, 1512–1524. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005, 95, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Sivori, S.; Vitale, M.; Morelli, L.; Sanseverino, L.; Augugliaro, R.; Bottino, C.; Moretta, L.; Moretta, A. P46, a Novel Natural Killer Cell-Specific Surface Molecule That Mediates Cell Activation. J. Exp. Med. 1997, 186, 1129–1136. [Google Scholar] [CrossRef] [Green Version]
- Pessino, A.; Sivori, S.; Bottino, C.; Malaspina, A.; Morelli, L.; Moretta, L.; Biassoni, R.; Moretta, A. Molecular cloning of NKp46: A novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998, 188, 953–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Billadeau, D.D.; Upshaw, J.L.; Schoon, R.A.; Dick, C.J.; Leibson, P.J. NKG2D-DAP10 triggers human NK cell-mediated killing via a Syk-independent regulatory pathway. Nat. Immunol. 2003, 4, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Gendzekhadze, K.; Norman, P.J.; Abi-Rached, L.; Graef, T.; Moesta, A.K.; Layrisse, Z.; Parham, P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc. Natl. Acad. Sci. USA 2009, 106, 18692–18697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bergen, J.; Thompson, A.; van der Slik, A.; Ottenhoff, T.H.M.; Gussekloo, J.; Koning, F. Phenotypic and functional characterization of CD4 T cells expressing killer Ig-like receptors. J. Immunol. 2004, 173, 6719–6726. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Pan, C.M.; Zhao, S.X.; Liang, J.; Gao, G.Q.; Zhang, X.M.; Yuan, G.Y.; Li, C.G.; Xue, L.Q.; Shen, M.; et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nat. Genet. 2011, 43, 897–901. [Google Scholar]
- Medici, M.; Porcu, E.; Pistis, G.; Teumer, A.; Brown, S.J.; Jensen, R.A.; Rawal, R.; Roef, G.L.; Plantinga, T.S.; Vermeulen, S.H.; et al. Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease. PLoS Genet. 2014, 10, e1004123. [Google Scholar] [CrossRef]
- Cho, W.K.; Jung, M.H.; Park, S.H.; Baek, I.C.; Choi, H.B.; Kim, T.G.; Suh, B.K. Association of MICA alleles with autoimmune thyroid disease in Korean children. Int. J. Endocrinol. 2012, 2012. [Google Scholar] [CrossRef]
- Kusś, A.; Szymański, K.; Peeters, R.P.; Mis’kiewicz, P.; Porcu, E.; Pistis, G.; Sanna, S.; Naitza, S.; Płoski, R.; Medici, M.; et al. The association of thyroid peroxidase antibody risk loci with susceptibility to and phenotype of Graves’ disease. Clin. Endocrinol. 2015, 83, 556–562. [Google Scholar] [CrossRef]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, M.M.; Ernst, J.; Wilder, S.P.; Kundaje, A.; Harris, R.S.; Libbrecht, M.; Giardine, B.; Ellenbogen, P.M.; Bilmes, J.A.; Birney, E.; et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013, 41, 827–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siu, C.; Wiseman, S.; Gakkhar, S.; Heravi-Moussavi, A.; Bilenky, M.; Carles, A.; Sierocinski, T.; Tam, A.; Zhao, E.; Kasaian, K.; et al. Characterization of the human thyroid epigenome. J. Endocrinol. 2017, 235, 153–165. [Google Scholar] [CrossRef]
- Martin, T.C.; Yet, I.; Tsai, P.-C.; Bell, J.T. coMET: Visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinform. 2015, 16, 131. [Google Scholar] [CrossRef] [Green Version]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Amino, N.; Yagawa, K.; Uemura, K.; Satoh, M.; Miyai, K.; Kumahara, Y. Association of serum antithyroid antibodies with lymphocytic infiltration of the thyroid gland: Studies of seventy autopsied cases. J. Clin. Endocrinol. Metab. 1978, 46, 859–862. [Google Scholar] [CrossRef] [PubMed]
- Kasagi, K.; Kousaka, T.; Higuchi, K.; Iida, Y.; Misaki, T.; Alam, M.S.; Miyamoto, S.; Yamabe, H.; Konishi, J. Clinical significance of measurements of antithyroid antibodies in the diagnosis of Hashimoto’s thyroiditis: Comparison with histological findings. Thyroid 1996, 6, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Carlé, A.; Laurberg, P.; Knudsen, N.; Perrild, H.; Ovesen, L.; Rasmussen, L.B.; Jorgensen, T.; Pedersen, I.B. Thyroid peroxidase and thyroglobulin auto-antibodies in patients with newly diagnosed overt hypothyroidism. Autoimmunity 2006, 39, 497–503. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.034226. [Google Scholar]
- Li, A.; Kim, J.; Smith, M.; Hughes, S.; Laderas, T. umapr. 2018. Available online: https://github.com/ropenscilabs/umapr (accessed on 12 February 2020).
- Nava, P.; Laukoetter, M.G.; Hopkins, A.M.; Laur, O.; Gerner-Smidt, K.; Green, K.J.; Parkos, C.A.; Nusrat, A. Desmoglein-2: A novel regulator of apoptosis in the intestinal epithelium. Mol. Biol. Cell 2007, 18, 4565–4578. [Google Scholar] [CrossRef] [Green Version]
- Effraimidis, G.; Wiersinga, W.M. Mechanisms in endocrinology: Autoimmune thyroid disease: Old and new players. Eur. J. Endocrinol. 2014, 170, R241–R252. [Google Scholar] [CrossRef] [PubMed]
- Rebuffat, S.A.; Nguyen, B.; Robert, B.; Castex, F.; Peraldi-Roux, S. Antithyroperoxidase antibody-dependent cytotoxicity in autoimmune thyroid disease. J. Clin. Endocrinol. Metab. 2008, 93, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Rebuffat, S.A.; Morin, M.; Nguyen, B.; Castex, F.; Robert, B.; Péraldi-Roux, S. Human recombinant anti-thyroperoxidase autoantibodies: In vitro cytotoxic activity on papillary thyroid cancer expressing TPO. Br. J. Cancer 2010, 102, 852–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalini, S.; Dorstyn, L.; Dawar, S.; Kumar, S. Old, new and emerging functions of caspases. Cell Death Differ. 2015, 22, 526–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berda-Haddad, Y.; Robert, S.; Salers, P.; Zekraoui, L.; Farnarier, C.; Dinarello, C.A.; Dignat-George, F.; Kaplanski, G. Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin-1alpha. Proc. Natl. Acad. Sci. USA 2011, 108, 20684–20689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grubeck-Loebenstein, B.; Buchan, G.; Chantry, D.; Kassal, H.; Londei, M.; Pirich, K.; Barrett, K.; Turner, M.; Waldhausl, W.; Feldmann, M. Analysis of intrathyroidal cytokine production in thyroid autoimmune disease: Thyroid follicular cells produce interleukin-1 alpha and interleukin-6. Clin. Exp. Immunol. 1989, 77, 324–330. [Google Scholar] [PubMed]
- Nilsson, M.; Husmark, J.; Björkman, U.; Ericson, L.E. Cytokines and thyroid epithelial integrity: Interleukin-1α induces dissociation of the junctional complex and paracellular leakage in filter- cultured human thyrocytes. J. Clin. Endocrinol. Metab. 1998, 83, 945–952. [Google Scholar] [CrossRef] [Green Version]
- Wagtmann, N.; Rajagopalan, S.; Winter, C.C.; Peruui, M.; Long, E.O. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity 1995, 3, 801–809. [Google Scholar] [CrossRef] [Green Version]
- Moesta, A.K.; Parham, P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front. Immunol. 2012, 3, 336. [Google Scholar] [CrossRef] [Green Version]
- Yamane-Ohnuki, N.; Kinoshita, S.; Inoue-Urakubo, M.; Kusunoki, M.; Iida, S.; Nakano, R.; Wakitani, M.; Niwa, R.; Sakurada, M.; Uchida, K.; et al. Establishment of FUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioeng. 2004, 87, 614–622. [Google Scholar] [CrossRef]
- Liu, Y.; Buil, A.; Collins, B.C.; Gillet, L.C.J.; Blum, L.C.; Cheng, L.-Y.; Vitek, O.; Mouritsen, J.; Lachance, G.; Spector, T.D.; et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 2015, 11, 786. [Google Scholar] [CrossRef] [PubMed]
- Ząbczyńska, M.; Polak, K.; Kozłowska, K.; Sokołowski, G.; Pocheć, E. The contribution of igg glycosylation to antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) in hashimoto’s thyroiditis: An in vitro model of thyroid autoimmunity. Biomolecules 2020, 10, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlers, M.; Thiel, A.; Bernecker, C.; Porwol, D.; Papewalis, C.; Willenberg, H.S.; Schinner, S.; Hautzel, H.; Scherbaum, W.A.; Schott, M. Evidence of a Combined Cytotoxic Thyroglobulin and Thyroperoxidase Epitope-Specific Cellular Immunity in Hashimoto’s Thyroiditis. J. Clin. Endocrinol. Metab. 2012, 97, 1347–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivier, E.; Nunès, J.A.; Vély, F. Natural killer cell signaling pathways. Science 2004, 306, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, E.; Koch, J.; Cerwenka, A.; Steinle, A. New prospects on the NKG2D/NKG2DL system for oncology. Oncoimmunology 2013, 2, e26097. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, T.C.; Ilieva, K.M.; Visconti, A.; Beaumont, M.; Kiddle, S.J.; Dobson, R.J.B.; Mangino, M.; Lim, E.M.; Pezer, M.; Steves, C.J.; et al. Dysregulated Antibody, Natural Killer Cell and Immune Mediator Profiles in Autoimmune Thyroid Diseases. Cells 2020, 9, 665. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030665
Martin TC, Ilieva KM, Visconti A, Beaumont M, Kiddle SJ, Dobson RJB, Mangino M, Lim EM, Pezer M, Steves CJ, et al. Dysregulated Antibody, Natural Killer Cell and Immune Mediator Profiles in Autoimmune Thyroid Diseases. Cells. 2020; 9(3):665. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030665
Chicago/Turabian StyleMartin, Tiphaine C., Kristina M. Ilieva, Alessia Visconti, Michelle Beaumont, Steven J. Kiddle, Richard J. B. Dobson, Massimo Mangino, Ee Mun Lim, Marija Pezer, Claire J. Steves, and et al. 2020. "Dysregulated Antibody, Natural Killer Cell and Immune Mediator Profiles in Autoimmune Thyroid Diseases" Cells 9, no. 3: 665. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030665