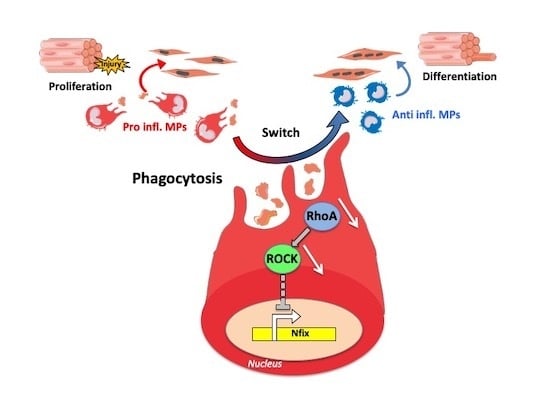
The Transcription Factor Nfix Requires RhoA-ROCK1 Dependent Phagocytosis to Mediate Macrophage Skewing during Skeletal Muscle Regeneration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Models and In Vivo Experimentations
2.2. Isolation of MPs from Skeletal Muscle
2.3. Histology and Immunofluorescence Analyses
2.4. Bone Marrow Derived MPs (BMDM) Culture
2.5. Myogenic Progenitor Cells (mpc) Culture
2.6. Phagocytosis Assay
2.7. Lentiviral Transduction
2.8. RNA Extraction and qRT-PCR
2.9. Protein Extraction and Western Blot
2.10. Image Acquisition and Quantification
2.11. Statistical Analysis
3. Results
3.1. Nfix is Expressed by Anti-Inflammatory MPs
3.2. Nfix Expression in MPs is Essential for Muscle Regeneration
3.3. Nfix is Required for MP Phenotypical Switch In Vivo and In Vitro
3.4. Nfix is Required for Macrophage Function on Mpcs In Vivo and In Vitro
3.5. Phagocytosis Induces the Expression of Nfix
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Duffield, J.S.; Forbes, S.J.; Constandinou, C.M.; Clay, S.; Partolina, M.; Vuthoori, S.; Wu, S.; Lang, R.; Iredale, J.P. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. Invest. 2005, 115, 56–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shechter, R.; Miller, O.; Yovel, G.; Rosenzweig, N.; London, A.; Ruckh, J.; Kim, K.W.; Klein, E.; Kalchenko, V.; Bendel, P.; et al. Recruitment of Beneficial M2 Macrophages to Injured Spinal Cord Is Orchestrated by Remote Brain Choroid Plexus. Immunity 2013, 38, 555–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, L.; Henry, A.; Poron, F.; Baba-Amer, Y.; Van Rooijen, N.; Plonquet, A.; Gherardi, R.K.; Chazaud, B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007, 204, 1057–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynn, T.A.; Vannella, K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef] [Green Version]
- Vannella, K.M.; Wynn, T.A. Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 2017, 79, 593–617. [Google Scholar] [CrossRef]
- Chazaud, B. Macrophages: Supportive cells for tissue repair and regeneration. Immunobiology 2014, 219, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Saclier, M.; Cuvellier, S.; Magnan, M.; Mounier, R.; Chazaud, B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013, 280, 4118–4130. [Google Scholar] [CrossRef]
- Mounier, R.; Théret, M.; Arnold, L.; Cuvellier, S.; Bultot, L.; Göransson, O.; Sanz, N.; Ferry, A.; Sakamoto, K.; Foretz, M.; et al. AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 2013, 18, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Perdiguero, E.; Sousa-Victor, P.; Ruiz-Bonilla, V.; Jardí, M.; Caelles, C.; Serrano, A.L.; Muñoz-Cánoves, P. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J. Cell Biol. 2011, 195, 307–322. [Google Scholar] [CrossRef] [Green Version]
- Lemke, G. How macrophages deal with death. Nat. Rev. Immunol. 2019, 19, 539–549. [Google Scholar] [CrossRef]
- Elliott, M.R.; Ravichandran, K.S. The Dynamics of Apoptotic Cell Clearance. Dev. Cell 2016, 38, 147–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.Q.; Freire-de-Lima, C.G.; Schiemann, W.P.; Bratton, D.L.; Vandivier, R.W.; Henson, P.M. Transcriptional and Translational Regulation of TGF-β Production in Response to Apoptotic Cells. J. Immunol. 2008, 181, 3575–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johann, A.M.; Barra, V.; Kuhn, A.M.; Weigert, A.; Von Knethen, A.; Brüne, B. Apoptotic cells induce arginase II in macrophages, thereby attenuating NO production. FASEB J. 2007, 21, 2704–2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochreiter-Hufford, A.; Ravichandran, K.S. Clearing the dead: Apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [Green Version]
- Freeman, S.A.; Grinstein, S. Phagocytosis: Receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014, 262, 193–215. [Google Scholar] [CrossRef]
- Tidball, J.G.; Wehling-Henricks, M. Damage and inflammation in muscular dystrophy: Potential implications and relationships with autoimmune myositis. Curr. Opin. Rheumatol. 2005, 17, 707–713. [Google Scholar] [CrossRef]
- Dort, J.; Fabre, P.; Molina, T.; Dumont, N.A. Macrophages Are Key Regulators of Stem Cells during Skeletal Muscle Regeneration and Diseases. Stem Cells Int. 2019, 2019. [Google Scholar] [CrossRef]
- Farup, J.; Madaro, L.; Puri, P.L.; Mikkelsen, U.R. Interactions between muscle stem cells, mesenchymal-derived cells and immune cells in muscle homeostasis, regeneration and disease. Cell Death Dis. 2015, 6, e1830. [Google Scholar] [CrossRef] [Green Version]
- Chazaud, B.; Brigitte, M.; Yacoub-Youssef, H.; Arnold, L.; Gherardi, R.; Sonnet, C.; Lafuste, P.; Chretien, F. Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc. Sport Sci. Rev. 2009, 37, 18–22. [Google Scholar] [CrossRef]
- Rigamonti, E.; Zordan, P.; Sciorati, C.; Rovere-Querini, P.; Brunelli, S. Macrophage plasticity in skeletal muscle repair. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Biressi, S.; Molinaro, M.; Cossu, G. Cellular heterogeneity during vertebrate skeletal muscle development. Dev. Biol. 2007, 308, 281–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauro, A. Satellite Cell of Skeletal Muscle Fibers. J. Biophys Biochem Cytol 1961, 9, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.C.; Rudnicki, M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar] [PubMed]
- Gronostajski, R.M. Roles of the NFI/CTF gene family in transcription and development. Gene 2000, 249, 31–45. [Google Scholar] [CrossRef]
- Messina, G.; Biressi, S.; Monteverde, S.; Magli, A.; Cassano, M.; Perani, L.; Roncaglia, E.; Tagliafico, E.; Starnes, L.; Campbell, C.E.; et al. Nfix Regulates Fetal-Specific Transcription in Developing Skeletal Muscle. Cell 2010, 140, 554–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pistocchi, A.; Gaudenzi, G.; Foglia, E.; Monteverde, S.; Moreno-Fortuny, A.; Pianca, A.; Cossu, G.; Cotelli, F.; Messina, G. Conserved and divergent functions of Nfix in skeletal muscle development during vertebrate evolution. Development 2013, 140, 2443. [Google Scholar] [CrossRef] [Green Version]
- Rossi, G.; Antonini, S.; Bonfanti, C.; Monteverde, S.; Vezzali, C.; Tajbakhsh, S.; Cossu, G.; Messina, G. Nfix Regulates Temporal Progression of Muscle Regeneration through Modulation of Myostatin Expression. Cell Rep. 2016, 14, 2238–2249. [Google Scholar] [CrossRef] [Green Version]
- Rossi, G.; Bonfanti, C.; Antonini, S.; Bastoni, M.; Monteverde, S.; Innocenzi, A.; Saclier, M.; Taglietti, V.; Messina, G. Silencing Nfix rescues muscular dystrophy by delaying muscle regeneration. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Saclier, M.; Yacoub-Youssef, H.; Mackey, A.L.; Arnold, L.; Ardjoune, H.; Magnan, M.; Sailhan, F.; Chelly, J.; Pavlath, G.K.; Mounier, R.; et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells 2013, 31, 384–396. [Google Scholar] [CrossRef] [Green Version]
- Wehling, M.; Spencer, M.J.; Tidball, J.G. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J. Cell Biol. 2001, 155, 123–131. [Google Scholar] [CrossRef]
- Kharraz, Y.; Guerra, J.; Mann, C.J.; Serrano, A.L.; Muñoz-Cánoves, P. Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediators Inflamm. 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Cánoves, P.; Serrano, A.L. Macrophages decide between regeneration and fibrosis in muscle. Trends Endocrinol. Metab. 2015, 26, 449–450. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.E.; Piper, M.; Plachez, C.; Yeh, Y.-T.; Baizer, J.S.; Osinski, J.M.; Litwack, E.D.; Richards, L.J.; Gronostajski, R.M. The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 2008, 8, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, T.; Mounier, R.; Horvath, A.; Cuvellier, S.; Dumont, F.; Poliska, S.; Ardjoune, H.; Juban, G.; Nagy, L.; Chazaud, B. Highly Dynamic Transcriptional Signature of Distinct Macrophage Subsets during Sterile Inflammation, Resolution, and Tissue Repair. J. Immunol. 2016, 196, 4771–4782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruffell, D.; Mourkioti, F.; Gambardella, A.; Kirstetter, P.; Lopez, R.G.; Rosenthal, N.; Nerlov, C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 2009, 106, 17475–17480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, M.; Liu, J.; Yang, Q.; Seok, H.Y.; Hu, X.; Deng, Z.-L.; Wang, D.-Z. MicroRNA-155 facilitates skeletal muscle regeneration by balancing pro- and anti-inflammatory macrophages. Cell Death Dis. 2016, 7, e2261. [Google Scholar] [CrossRef]
- Saclier, M.; Theret, M.; Mounier, R.; Chazaud, B. Effects of macrophage conditioned-medium on murine and human muscle cells: Analysis of proliferation, differentiation, and fusion. Proc. Natl. Acad. Sci. USA 2009, 106, 17475–17480. [Google Scholar]
- Arnold, L.; Perrin, H.; de Chanville, C.B.; Saclier, M.; Hermand, P.; Poupel, L.; Guyon, E.; Licata, F.; Carpentier, W.; Vilar, J.; et al. CX3CR1 deficiency promotes muscle repair and regeneration by enhancing macrophage ApoE production. Nat. Commun. 2015, 6, 8972. [Google Scholar] [CrossRef] [Green Version]
- Taglietti, V.; Angelini, G.; Mura, G.; Bonfanti, C.; Caruso, E.; Monteverde, S.; Le Carrou, G.; Tajbakhsh, S.; Relaix, F.; Messina, G. RhoA and ERK signalling regulate the expression of the transcription factor Nfix in myogenic cells. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Bros, M.; Haas, K.; Moll, L. Grabbe RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells 2019, 8, 733. [Google Scholar] [CrossRef] [Green Version]
- Nakaya, M.; Tanaka, M.; Okabe, Y.; Hanayama, R.; Nagata, S. Opposite effects of Rho family GTPases on engulfment of apoptotic cells by macrophages. J. Biol. Chem. 2006, 281, 8836–8842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Königs, V.; Jennings, R.; Vogl, T.; Horsthemke, M.; Bachg, A.C.; Xu, Y.; Grobe, K.; Brakebusch, C.; Schwab, A.; Bähler, M.; et al. Mouse Macrophages completely lacking Rho subfamily GTPases (RhoA, RhoB, and RhoC) have severe lamellipodial retraction defects, but robust chemotactic navigation and altered motility. J. Biol. Chem. 2014, 289, 30772–30784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Kim, S.; Bae, D.J.; Park, S.Y.; Lee, G.Y.; Park, G.M.; Kim, I.S. Coordinated balance of Rac1 and RhoA plays key roles in determining phagocytic appetite. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.; Zalucki, O.; Gobius, I.; McDonald, H.; Osinki, J.; Harvey, T.J.; Essebier, A.; Vidovic, D.; Gladwyn-Ng, I.; Burne, T.H.; et al. Transcriptional regulation of intermediate progenitor cell generation during hippocampal development. Development 2016, 143, 4620–4630. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.; Dixon, C.; Cato, K.; Heng, Y.H.E.; Kurniawan, N.D.; Ullmann, J.F.P.; Janke, A.L.; Gronostajski, R.M.; Richards, L.J.; Burne, T.H.J.; et al. Heterozygosity for Nuclear Factor One X Affects Hippocampal-Dependent Behaviour in Mice. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.; Essebier, A.; Gronostajski, R.M.; Boden, M.; Wainwright, B.J.; Harvey, T.J.; Piper, M. Cell-type-specific expression of NFIX in the developing and adult cerebellum. Brain Struct. Funct. 2017, 222, 2251–2270. [Google Scholar] [CrossRef]
- Holmfeldt, P.; Pardieck, J.; Saulsberry, A.C.; Nandakumar, S.K.; Finkelstein, D.; Gray, J.T.; Persons, D.A.; Mckinney-freeman, S. Nfix is a novel regulator of murine hematopoietic stem and progenitor cell survival. Blood 2015, 122, 2987–2997. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, C.; Campos, J.; Osinski, J.M.; Gronostajski, R.M.; Michie, A.M.; Keeshan, K. Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS ONE 2015, 10, 1–15. [Google Scholar] [CrossRef]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; de Bruijn, M.; Rodewald, H.R.; Geissmann, F. The Origin of Tissue-Resident Macrophages: When an Erythro-myeloid Progenitor Is an Erythro-myeloid Progenitor. Immunity 2015, 43, 1023–1024. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Kim, I.S. Engulfment signals and the phagocytic machinery for apoptotic cell clearance. Exp. Mol. Med. 2017, 49. [Google Scholar] [CrossRef] [Green Version]
- Haney, M.S.; Bohlen, C.J.; Morgens, D.W.; Ousey, J.A.; Barkal, A.A.; Tsui, C.K.; Ego, B.K.; Levin, R.; Kamber, R.A.; Collins, H.; et al. Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat. Genet. 2018, 50, 1716–1727. [Google Scholar] [CrossRef] [PubMed]
- Ngambenjawong, C.; Gustafson, H.H.; Pun, S.H. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv. Drug Deliv. Rev. 2017, 114, 206–221. [Google Scholar] [CrossRef] [Green Version]
- Tang, P.M.K.; Nikolic-Paterson, D.J.; Lan, H.Y. Macrophages: Versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 2019, 15, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Lemos, D.R.; Babaeijandaghi, F.; Low, M.; Chang, C.-K.; Lee, S.T.; Fiore, D.; Zhang, R.-H.; Natarajan, A.; Nedospasov, S.A.; Rossi, F.M. V Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med. 2015, 21, 786–794. [Google Scholar] [CrossRef]
- Ueha, S.; Shand, F.H.W.; Matsushima, K. Cellular and molecular mechanisms of chronic inflammation-associated organ fibrosis. Front. Immunol. 2012, 3, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Tan, R.J.; Liu, Y. Macrophage-derived TGF-beta in renal fibrosis: Not a macro- impact after all. Am. J. Physiol. Renal Physiol. 2013, 1–7. [Google Scholar]
- Mann, C.J.; Perdiguero, E.; Kharraz, Y.; Aguilar, S.; Pessina, P.; Serrano, A.L.; Muñoz-Cánoves, P. Aberrant repair and fibrosis development in skeletal muscle. Skelet. Muscle 2011, 1, 21. [Google Scholar] [CrossRef] [Green Version]
- Vidal, B.; Serrano, A.L.; Tjwa, M.; Suelves, M.; Ardite, E.; De Mori, R.; Baeza-Raja, B.; De Lagrán, M.M.; Lafuste, P.; Ruiz-Bonilla, V.; et al. Fibrinogen drives dystrophic muscle fibrosis via a TGFβ/alternative macrophage activation pathway. Genes Dev. 2008, 22, 1747–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tidball, J.G.; Wehling-Henricks, M. Shifts in macrophage cytokine production drive muscle fibrosis. Nat. Med. 2015, 21, 665–666. [Google Scholar] [CrossRef]
- Pakshir, P.; Hinz, B. The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 2018, 68–69, 81–93. [Google Scholar] [CrossRef]
- Smith, L.R.; Barton, E.R. Regulation of fibrosis in muscular dystrophy. Matrix Biol. 2018, 68–69, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Biernacka, A.; Dobaczewski, M.; Frangogiannis, N.G. TGF-β signaling in fibrosis. Growth Factors 2011, 29, 196–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saclier, M.; Lapi, M.; Bonfanti, C.; Rossi, G.; Antonini, S.; Messina, G. The Transcription Factor Nfix Requires RhoA-ROCK1 Dependent Phagocytosis to Mediate Macrophage Skewing during Skeletal Muscle Regeneration. Cells 2020, 9, 708. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030708
Saclier M, Lapi M, Bonfanti C, Rossi G, Antonini S, Messina G. The Transcription Factor Nfix Requires RhoA-ROCK1 Dependent Phagocytosis to Mediate Macrophage Skewing during Skeletal Muscle Regeneration. Cells. 2020; 9(3):708. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030708
Chicago/Turabian StyleSaclier, Marielle, Michela Lapi, Chiara Bonfanti, Giuliana Rossi, Stefania Antonini, and Graziella Messina. 2020. "The Transcription Factor Nfix Requires RhoA-ROCK1 Dependent Phagocytosis to Mediate Macrophage Skewing during Skeletal Muscle Regeneration" Cells 9, no. 3: 708. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030708