Modulation of mTORC1 Signaling Pathway by HIV-1
Abstract
:1. Introduction
2. mTORC1, a Main Metabolic Network of the Cell
3. mTORC1 in the Immune System
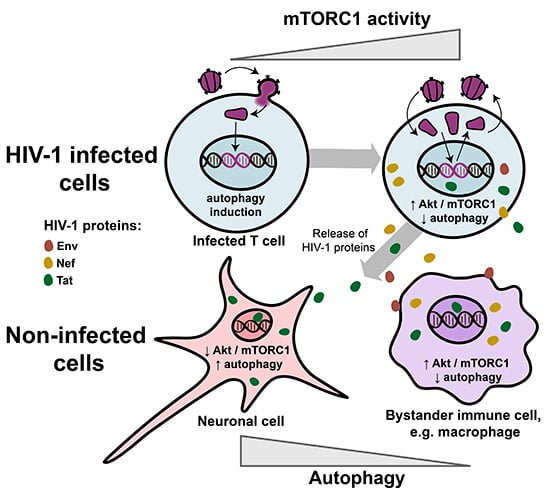
4. HIV-1 Modulates mTORC1 Activity
5. Regulation of Autophagy by HIV-1
6. mTORC1 in HIV-1 Latency
7. mTORC1 Pathway in HIV-1-Related Diseases
8. Concluding Remarks
9. Outstanding Questions
Funding
Conflicts of Interest
References
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How Viral and Intracellular Bacterial Pathogens Reprogram the Metabolism of Host Cells to Allow Their Intracellular Replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrbod, P.; Ande, S.R.; Alizadeh, J.; Rahimizadeh, S.; Shariati, A.; Malek, H.; Hashemi, M.; Glover, K.K.M.; Sher, A.A.; Coombs, K.M.; et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 2019, 10, 376–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, L.; Mostowy, S.; Sancho-Shimizu, V. Autophagy-Virus Interplay: From Cell Biology to Human Disease. Front. Cell Dev. Biol. 2018, 6, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Sage, V.; Cinti, A.; Amorim, R.; Mouland, A. Adapting the Stress Response: Viral Subversion of the mTOR Signaling Pathway. Viruses 2016, 8, 152. [Google Scholar] [CrossRef]
- Votteler, J.; Schubert, U. Human Immunodeficiency Viruses: Molecular Biology. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2008; pp. 517–525. [Google Scholar]
- Burdick, R.C.; Li, C.; Munshi, M.H.; Rawson, J.M.O.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Ma, Y.; Vassetzky, Y.; Dokudovskaya, S. mTORC1 pathway in DNA damage response. Biochim. Biophys. Acta—Mol. Cell Res. 2018, 1865, 1293–1311. [Google Scholar] [CrossRef]
- Cervera, C.; Cofan, F.; Hernandez, C.; Soy, D.; Marcos, M.A.; Sanclemente, G.; Bodro, M.; Moreno, A.; Diekmann, F.; Campistol, J.M.; et al. Effect of mammalian target of rapamycin inhibitors on cytomegalovirus infection in kidney transplant recipients receiving polyclonal antilymphocyte globulins: A propensity score-matching analysis. Transpl. Int. 2016, 29, 1216–1225. [Google Scholar] [CrossRef]
- Piñana, J.L.; Perez-Pitarch, A.; Guglieri-Lopez, B.; Giménez, E.; Hernandez-Boluda, J.C.; Terol, M.J.; Ferriols-Lisart, R.; Solano, C.; Navarro, D. Sirolimus exposure and the occurrence of cytomegalovirus DNAemia after allogeneic hematopoietic stem cell transplantation. Am. J. Transplant. 2018, 18, 2885–2894. [Google Scholar] [CrossRef] [Green Version]
- Mannick, J.B.; Morris, M.; Hockey, H.-U.P.; Roma, G.; Beibel, M.; Kulmatycki, K.; Watkins, M.; Shavlakadze, T.; Zhou, W.; Quinn, D.; et al. TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci. Transl. Med. 2018, 10, eaaq1564. [Google Scholar] [CrossRef] [Green Version]
- Heredia, A.; Amoroso, A.; Davis, C.; Le, N.; Reardon, E.; Dominique, J.K.; Klingebiel, E.; Gallo, R.C.; Redfield, R.R. Rapamycin causes down-regulation of CCR5 and accumulation of anti-HIV -chemokines: An approach to suppress R5 strains of HIV-1. Proc. Natl. Acad. Sci. USA 2003, 100, 10411–10416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donia, M.; McCubrey, J.A.; Bendtzen, K.; Nicoletti, F. Potential use of rapamycin in HIV infection. Br. J. Clin. Pharmacol. 2010, 70, 784–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicoletti, F.; Fagone, P.; Meroni, P.; McCubrey, J.; Bendtzen, K. mTOR as a multifunctional therapeutic target in HIV infection. Drug Discov. Today 2011, 16, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Heredia, A.; Le, N.; Gartenhaus, R.B.; Sausville, E.; Medina-Moreno, S.; Zapata, J.C.; Davis, C.; Gallo, R.C.; Redfield, R.R. Targeting of mTOR catalytic site inhibits multiple steps of the HIV-1 lifecycle and suppresses HIV-1 viremia in humanized mice. Proc. Natl. Acad. Sci. USA 2015, 112, 9412–9417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [Green Version]
- Dibble, C.C.; Elis, W.; Menon, S.; Qin, W.; Klekota, J.; Asara, J.M.; Finan, P.M.; Kwiatkowski, D.J.; Murphy, L.O.; Manning, B.D. TBC1D7 Is a Third Subunit of the TSC1-TSC2 Complex Upstream of mTORC1. Mol. Cell 2012, 47, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Vander Haar, E.; Lee, S.; Bandhakavi, S.; Griffin, T.J.; Kim, D.-H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323. [Google Scholar] [CrossRef]
- Dokudovskaya, S.; Rout, M.P. SEA you later alli-GATOR--a dynamic regulator of the TORC1 stress response pathway. J. Cell Sci. 2015, 128, 2219–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfson, R.L.; Sabatini, D.M. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 2017, 26, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Howell, J.J.; Hellberg, K.; Turner, M.; Talbott, G.; Kolar, M.J.; Ross, D.S.; Hoxhaj, G.; Saghatelian, A.; Shaw, R.J.; Manning, B.D. Metformin Inhibits Hepatic mTORC1 Signaling via Dose-Dependent Mechanisms Involving AMPK and the TSC Complex. Cell Metab. 2017, 25, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Brugarolas, J.; Lei, K.; Hurley, R.L.; Manning, B.D.; Reiling, J.H.; Hafen, E.; Witters, L.A.; Ellisen, L.W.; Kaelin, W.G. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004, 18, 2893–2904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awasthi, P.; Foiani, M.; Kumar, A. ATM and ATR signaling at a glance. J. Cell Sci. 2016, 129, 1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montaner, S.; Sodhi, A.; Pece, S.; Mesri, E.A.; Gutkind, J.S. The Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor promotes endothelial cell survival through the activation of Akt/protein kinase B. Cancer Res. 2001, 61, 2641–2648. [Google Scholar]
- Powell, J.D.; Pollizzi, K.N.; Heikamp, E.B.; Horton, M.R. Regulation of Immune Responses by mTOR. Annu. Rev. Immunol. 2012, 30, 39–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linke, M.; Fritsch, S.D.; Sukhbaatar, N.; Hengstschläger, M.; Weichhart, T. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett. 2017, 591, 3089–3103. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.N.; Ramírez-Komo, J.A.; Park, H.; Iritani, B.M. Title: Control of B Lymphocyte Development and Functions by the mTOR Signaling Pathways. Cytokine Growth Factor Rev. 2017, 35, 47–62. [Google Scholar] [CrossRef]
- Liu, C.; Chapman, N.M.; Karmaus, P.W.F.; Zeng, H.; Chi, H. mTOR and metabolic regulation of conventional and regulatory T cells. J. Leukoc. Biol. 2015, 97, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Weichhart, T.; Hengstschläger, M.; Linke, M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015, 15, 599–614. [Google Scholar] [CrossRef]
- Delgoffe, G.M.; Kole, T.P.; Zheng, Y.; Zarek, P.E.; Matthews, K.L.; Xiao, B.; Worley, P.F.; Kozma, S.C.; Powell, J.D. The mTOR Kinase Differentially Regulates Effector and Regulatory T Cell Lineage Commitment. Immunity 2009, 30, 832–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgoffe, G.M.; Pollizzi, K.N.; Waickman, A.T.; Heikamp, E.; Meyers, D.J.; Horton, M.R.; Xiao, B.; Worley, P.F.; Powell, J.D. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 2011, 12, 295–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurebayashi, Y.; Nagai, S.; Ikejiri, A.; Ohtani, M.; Ichiyama, K.; Baba, Y.; Yamada, T.; Egami, S.; Hoshii, T.; Hirao, A.; et al. PI3K-Akt-mTORC1-S6K1/2 Axis Controls Th17 Differentiation by Regulating Gfi1 Expression and Nuclear Translocation of RORγ. Cell Rep. 2012, 1, 360–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Shrestha, S.; Zeng, H.; Karmaus, P.W.F.; Neale, G.; Vogel, P.; Guertin, D.A.; Lamb, R.F.; Chi, H. T Cell Exit from Quiescence and Differentiation into Th2 Cells Depend on Raptor-mTORC1-Mediated Metabolic Reprogramming. Immunity 2013, 39, 1043–1056. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Jin, H.-S.; Lopez, J.; Elly, C.; Kim, G.; Murai, M.; Kronenberg, M.; Liu, Y.-C. TSC1 regulates the balance between effector and regulatory T cells. J. Clin. Investig. 2013, 123, 5165–5178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, L.V.; Rolf, J.; Emslie, E.; Shi, Y.-B.; Taylor, P.M.; Cantrell, D.A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013, 14, 500–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakaya, M.; Xiao, Y.; Zhou, X.; Chang, J.-H.; Chang, M.; Cheng, X.; Blonska, M.; Lin, X.; Sun, S.-C. Inflammatory T Cell Responses Rely on Amino Acid Transporter ASCT2 Facilitation of Glutamine Uptake and mTORC1 Kinase Activation. Immunity 2014, 40, 692–705. [Google Scholar] [CrossRef] [Green Version]
- Araki, K.; Turner, A.P.; Shaffer, V.O.; Gangappa, S.; Keller, S.A.; Bachmann, M.F.; Larsen, C.P.; Ahmed, R. mTOR regulates memory CD8 T-cell differentiation. Nature 2009, 460, 108–112. [Google Scholar] [CrossRef] [Green Version]
- Pearce, E.L.; Walsh, M.C.; Cejas, P.J.; Harms, G.M.; Shen, H.; Wang, L.-S.; Jones, R.G.; Choi, Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 2009, 460, 103–107. [Google Scholar] [CrossRef]
- Rao, R.R.; Li, Q.; Odunsi, K.; Shrikant, P.A. The mTOR Kinase Determines Effector versus Memory CD8 + T Cell Fate by Regulating the Expression of Transcription Factors T-bet and Eomesodermin. Immunity 2010, 32, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Yang, K.; Cloer, C.; Neale, G.; Vogel, P.; Chi, H. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 2013, 499, 485–490. [Google Scholar] [CrossRef] [Green Version]
- De Rosa, V.; Galgani, M.; Porcellini, A.; Colamatteo, A.; Santopaolo, M.; Zuchegna, C.; Romano, A.; De Simone, S.; Procaccini, C.; La Rocca, C.; et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 2015, 16, 1174–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerriets, V.A.; Kishton, R.J.; Johnson, M.O.; Cohen, S.; Siska, P.J.; Nichols, A.G.; Warmoes, M.O.; de Cubas, A.A.; MacIver, N.J.; Locasale, J.W.; et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 2016, 17, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Long, L.; Yang, K.; Guy, C.; Shrestha, S.; Chen, Z.; Wu, C.; Vogel, P.; Neale, G.; Green, D.R.; et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 2016, 17, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, T.N.; Ramírez, J.A.; Tsang, M.; Park, H.; Margineantu, D.H.; Hockenbery, D.M.; Iritani, B.M. Conditional Disruption of Raptor Reveals an Essential Role for mTORC1 in B Cell Development, Survival, and Metabolism. J. Immunol. 2016, 197, 2250–2260. [Google Scholar] [CrossRef] [Green Version]
- Raybuck, A.L.; Cho, S.H.; Li, J.; Rogers, M.C.; Lee, K.; Williams, C.L.; Shlomchik, M.; Thomas, J.W.; Chen, J.; Williams, J.V.; et al. B Cell–Intrinsic mTORC1 Promotes Germinal Center–Defining Transcription Factor Gene Expression, Somatic Hypermutation, and Memory B Cell Generation in Humoral Immunity. J. Immunol. 2018, 200, 2627–2639. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Pruitt, M.; Tran, D.; Du Bois, W.; Zhang, K.; Patel, R.; Hoover, S.; Simpson, R.M.; Simmons, J.; Gary, J.; et al. B Cell–Specific Deficiencies in mTOR Limit Humoral Immune Responses. J. Immunol. 2013, 191, 1692–1703. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.S.; Zhang, Z.-G.; Zhou, Z.; Du, G.-Y.; Li, H.; Yu, X.-Y.; Huang, Y.-H. PKM2-mediated inhibition of autophagy facilitates Tat’s inducing HIV-1 transactivation. Arch. Biochem. Biophys. 2017, 625–626, 17–23. [Google Scholar] [CrossRef]
- Kumar, B.; Arora, S.; Ahmed, S.; Banerjea, A.C. Hyperactivation of mammalian target of rapamycin complex 1 by HIV-1 is necessary for virion production and latent viral reactivation. FASEB J. 2017, 31, 180–191. [Google Scholar] [CrossRef] [Green Version]
- Van Grol, J.; Subauste, C.; Andrade, R.M.; Fujinaga, K.; Nelson, J.; Subauste, C.S. HIV-1 inhibits autophagy in bystander macrophage/ monocytic cells through Src-Akt and STAT3. PLoS ONE 2010, 5, e11733. [Google Scholar] [CrossRef] [Green Version]
- Xue, M.; Yao, S.; Hu, M.; Li, W.; Hao, T.; Zhou, F.; Zhu, X.; Lu, H.; Qin, D.; Yan, Q.; et al. HIV-1 Nef and KSHV oncogene K1 synergistically promote angiogenesis by inducing cellular miR-718 to regulate the PTEN/AKT/mTOR signaling pathway. Nucleic Acids Res. 2014, 42, 9862–9879. [Google Scholar] [CrossRef]
- Cinti, A.; Le Sage, V.; Milev, M.P.; Valiente-Echeverría, F.; Crossie, C.; Miron, M.-J.; Panté, N.; Olivier, M.; Mouland, A.J. HIV-1 enhances mTORC1 activity and repositions lysosomes to the periphery by co-opting Rag GTPases. Sci. Rep. 2017, 7, 5515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planas, D.; Routy, J.; Ancuta, P.; Planas, D.; Zhang, Y.; Monteiro, P.; Goulet, J.; Gosselin, A. HIV-1 selectively targets gut-homing mechanisms Find the latest version: HIV-1 selectively targets gut-homing mechanisms. JCI Insight 2017, 2, e93230. [Google Scholar] [CrossRef] [PubMed]
- Molle, D.; Segura-Morales, C.; Camus, G.; Berlioz-Torrent, C.; Kjems, J.; Basyuk, E.; Bertrand, E. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J. Biol. Chem. 2009, 284, 19727–19743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvez, M.; Hseeh, G.; Benzer, S.; Brown, A.M. Osteopontin counters human immunodeficiency virus type 1–induced impairment of neurite growth through mammalian target of rapamycin and beta-integrin signaling pathways. J. Neurovirol. 2019, 25, 384–396. [Google Scholar] [CrossRef] [Green Version]
- Blanchet, F.P.; Moris, A.; Nikolic, D.S.; Lehmann, M.; Cardinaud, S.; Stalder, R.; Garcia, E.; Dinkins, C.; Leuba, F.; Wu, L.; et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 2010, 32, 654–669. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, W.; Tong, P.; Leung, C.-K.; Yang, G.; Li, Z.; Li, N.; Sun, X.; Han, Y.; Lu, C.; et al. Autophagy Induction by HIV-Tat and Methamphetamine in Primary Midbrain Neuronal Cells of Tree Shrews via the mTOR Signaling and ATG5/ATG7 Pathway. Front. Neurosci. 2018, 12, 921. [Google Scholar] [CrossRef]
- Poggi, A.; Carosio, R.; Fenoglio, D.; Brenci, S.; Murdaca, G.; Setti, M.; Indiveri, F.; Scabini, S.; Ferrero, E.; Zocchi, M.R. Migration of V delta 1 and V delta 2 T cells in response to CXCR3 and CXCR4 ligands in healthy donors and HIV-1-infected patients: Competition by HIV-1 Tat. Blood 2004, 103, 2205–2213. [Google Scholar] [CrossRef]
- Germini, D.; Tsfasman, T.; Klibi, M.; El-Amine, R.; Pichugin, A.; Iarovaia, O.V.; Bilhou-Nabera, C.; Subra, F.; Bou Saada, Y.; Sukhanova, A.; et al. HIV Tat induces a prolonged MYC relocalization next to IGH in circulating B-cells. Leukemia 2017, 31, 2515–2522. [Google Scholar] [CrossRef]
- Kurnaeva, M.A.; Sheval, E.V.; Musinova, Y.R.; Vassetzky, Y.S. Tat basic domain: A “Swiss army knife” of HIV-1 Tat? Rev. Med. Virol. 2019, 29, e2031. [Google Scholar] [CrossRef]
- Lahiri, V.; Hawkins, W.D.; Klionsky, D.J. Watch What You (Self-) Eat: Autophagic Mechanisms that Modulate Metabolism. Cell Metab. 2019, 29, 803–826. [Google Scholar] [CrossRef] [Green Version]
- Dossou, A.S.; Basu, A. The Emerging Roles of mTORC1 in Macromanaging Autophagy. Cancers (Basel) 2019, 11, 1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Beek, N.; Klionsky, D.J.; Reggiori, F. Genetic aberrations in macroautophagy genes leading to diseases. Biochim. Biophys. Acta—Mol. Cell Res. 2018, 1865, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Nardacci, R.; Ciccosanti, F.; Marsella, C.; Ippolito, G.; Piacentini, M.; Fimia, G.M. Role of autophagy in HIV infection and pathogenesis. J. Intern. Med. 2017, 281, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, Y.; Torresilla, C.; Rassart, É.; Barbeau, B. Implication of Different HIV-1 Genes in the Modulation of Autophagy. Viruses 2017, 9, 389. [Google Scholar] [CrossRef] [Green Version]
- Kyei, G.B.; Dinkins, C.; Davis, A.S.; Roberts, E.; Singh, S.B.; Dong, C.; Wu, L.; Kominami, E.; Ueno, T.; Yamamoto, A.; et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009, 186, 255–268. [Google Scholar] [CrossRef]
- Campbell, G.R.; Rawat, P.; Bruckman, R.S.; Spector, S.A. Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration. PLoS Pathog. 2015, 11, e1005018. [Google Scholar] [CrossRef] [Green Version]
- Castro-Gonzalez, S.; Shi, Y.; Colomer-Lluch, M.; Song, Y.; Mowery, K.; Almodovar, S.; Bansal, A.; Kirchhoff, F.; Sparrer, K.; Liang, C.; et al. HIV-1 Nef counteracts autophagy restriction by enhancing the association between BECN1 and its inhibitor BCL2 in a PRKN-dependent manner. Autophagy 2020, 8627, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.; Young, L.N.; Morris, K.L.; von Bülow, S.; Schöneberg, J.; Yamamoto-Imoto, H.; Oe, Y.; Yamamoto, K.; Nakamura, S.; Stjepanovic, G.; et al. Bidirectional Control of Autophagy by BECN1 BARA Domain Dynamics. Mol. Cell 2019, 73, 339–353. [Google Scholar] [CrossRef] [Green Version]
- Espert, L.; Denizot, M.; Grimaldi, M.; Robert-Hebmann, V.; Gay, B.; Varbanov, M.; Codogno, P.; Biard-Piechaczyk, M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Investig. 2006, 116, 2161–2172. [Google Scholar] [CrossRef]
- Espert, L.; Varbanov, M.; Robert-Hebmann, V.; Sagnier, S.; Robbins, I.; Sanchez, F.; Lafont, V.; Biard-Piechaczyk, M. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS ONE 2009, 4, e5787. [Google Scholar] [CrossRef]
- Sagnier, S.; Daussy, C.F.; Borel, S.; Robert-Hebmann, V.; Faure, M.; Blanchet, F.P.; Beaumelle, B.; Biard-Piechaczyk, M.; Espert, L. Autophagy Restricts HIV-1 Infection by Selectively Degrading Tat in CD4 + T Lymphocytes. J. Virol. 2015, 89, 615–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, G.R.; Bruckman, R.S.; Herns, S.D.; Joshi, S.; Durden, D.L.; Spector, S.A. Induction of autophagy by PI3K/MTOR and PI3K/MTOR/BRD4 inhibitors suppresses HIV-1 replication. J. Biol. Chem. 2018, 293, 5808–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaisal, J.; Machado, A.; Galais, M.; Robert-Hebmann, V.; Arnauné-Pelloquin, L.; Espert, L.; Biard-Piechaczyk, M. HIV-1 Vpr inhibits autophagy during the early steps of infection of CD4 T cells. Biol. Cell 2019, 111, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Borel, S.; Robert-Hebmann, V.; Alfaisal, J.; Jain, A.; Faure, M.; Espert, L.; Chaloin, L.; Paillart, J.-C.; Johansen, T.; Biard-Piechaczyk, M. HIV-1 viral infectivity factor interacts with microtubule-associated protein light chain 3 and inhibits autophagy. AIDS 2015, 29, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Yakasai, A.M. Impact of Symptomatic HIV- Related Neurocognitive Disorders in Survival of HIV- Infected Individuals: A Systematic Review and Meta-Analyses. J. Neuroinfect. Dis. 2015, 06, 1–7. [Google Scholar] [CrossRef]
- Fields, J.; Dumaop, W.; Elueteri, S.; Campos, S.; Serger, E.; Trejo, M.; Kosberg, K.; Adame, A.; Spencer, B.; Rockenstein, E.; et al. HIV-1 Tat Alters Neuronal Autophagy by Modulating Autophagosome Fusion to the Lysosome: Implications for HIV-Associated Neurocognitive Disorders. J. Neurosci. 2015, 35, 1921–1938. [Google Scholar] [CrossRef] [Green Version]
- Bruno, A.P.; De Simone, F.I.; Iorio, V.; De Marco, M.; Khalili, K.; Sariyer, I.K.; Capunzo, M.; Nori, S.L.; Rosati, A. HIV-1 Tat protein induces glial cell autophagy through enhancement of BAG3 protein levels. Cell Cycle 2014, 13, 3640–3644. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Dong, H.; Ye, X.; Zhong, L.; Cao, T.; Xu, Q.; Wang, J.; Zhang, Y.; Xu, J.; Wang, W.; et al. HIV-1 Tat increases BAG3 via NF-κB signaling to induce autophagy during HIV-associated neurocognitive disorder. Cell Cycle 2018, 17, 1614–1623. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.F.; Li, Q.; Li, J.; Wong, N.; Li, Z.; Huang, J.; Yang, G.; Sham, P.C.; Li, S.B.; Lu, G. HIV-1 Tat and methamphetamine co-induced oxidative cellular injury is mitigated by N-acetylcysteine amide (NACA) through rectifying mTOR signaling. Toxicol. Lett. 2018, 299, 159–171. [Google Scholar] [CrossRef]
- Qi, L.; Gang, L.; Hang, K.W.; Ling, C.H.; Xiaofeng, Z.; Zhen, L.; David Wai, Y.; Sang, P.W. Programmed neuronal cell death induced by HIV-1 tat and methamphetamine. Microsc. Res. Tech. 2011, 74, 1139–1144. [Google Scholar] [CrossRef]
- Thangaraj, A.; Periyasamy, P.; Liao, K.; Bendi, V.S.; Callen, S.; Pendyala, G.; Buch, S. HIV-1 TAT-mediated microglial activation: Role of mitochondrial dysfunction and defective mitophagy. Autophagy 2018, 14, 1596–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sereti, I.; Krebs, S.J.; Phanuphak, N.; Fletcher, J.L.; Slike, B.; Pinyakorn, S.; O’Connell, R.J.; Rupert, A.; Chomont, N.; Valcour, V.; et al. Persistent, Albeit Reduced, Chronic Inflammation in Persons Starting Antiretroviral Therapy in Acute HIV Infection. Clin. Infect. Dis. 2017, 64, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Guardigni, V.; Pencina, K.M.; Amato, A.A.; Floyd, M.; Brawley, B.; Mozeleski, B.; McKinnon, J.; Woodbury, E.; Heckel, E.; et al. Atypical Skeletal Muscle Profiles in Human Immunodeficiency Virus-Infected Asymptomatic Middle-Aged Adults. Clin. Infect. Dis. 2018, 66, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Thrift, A.P.; Chiao, E.Y. Are Non-HIV Malignancies Increased in the HIV-Infected Population? Curr. Infect. Dis. Rep. 2018, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Shmakova, A.; Germini, D.; Vassetzky, Y. HIV-1, HAART and cancer: A complex relationship. Int. J. Cancer 2020, 146, 2666–2679. [Google Scholar] [CrossRef] [PubMed]
- Hamers, R.L.; Rinke de Wit, T.F.; Holmes, C.B. HIV drug resistance in low-income and middle-income countries. Lancet HIV 2018, 5, e588–e596. [Google Scholar] [CrossRef]
- Chimukangara, B.; Lessells, R.J.; Rhee, S.Y.; Giandhari, J.; Kharsany, A.B.M.; Naidoo, K.; Lewis, L.; Cawood, C.; Khanyile, D.; Ayalew, K.A.; et al. Trends in Pretreatment HIV-1 Drug Resistance in Antiretroviral Therapy-naive Adults in South Africa, 2000–2016: A Pooled Sequence Analysis. EClinicalMedicine 2019, 9, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Blankson, J.N.; Persaud, D.; Siliciano, R.F. The Challenge of Viral Reservoirs in HIV-1 Infection. Annu. Rev. Med. 2002, 53, 557–593. [Google Scholar] [CrossRef]
- Eisele, E.; Siliciano, R.F. Redefining the Viral Reservoirs that Prevent HIV-1 Eradication. Immunity 2012, 37, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Vanhamel, J.; Bruggemans, A.; Debyser, Z. Establishment of latent HIV-1 reservoirs: What do we really know? J. Virus Erad. 2019, 5, 3–9. [Google Scholar]
- Abner, E.; Jordan, A. HIV “shock and kill” therapy: In need of revision. Antivir. Res. 2019, 166, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, M.M.; Tang, Y.; Li, D.; Jiang, G. Deep latency: A new insight into a functional HIV cure. EBioMedicine 2019, 45, 624–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spivak, A.M.; Planelles, V. Novel Latency Reversal Agents for HIV-1 Cure. Annu. Rev. Med. 2018, 69, 421–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besnard, E.; Hakre, S.; Kampmann, M.; Lim, H.W.; Hosmane, N.N.; Martin, A.; Bassik, M.C.; Verschueren, E.; Battivelli, E.; Chan, J.; et al. The mTOR Complex Controls HIV Latency. Cell Host Microbe 2016, 20, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.R.; Pollack, R.A.; Capoferri, A.; Ambinder, R.F.; Durand, C.M.; Siliciano, R.F. Rapamycin-mediated mTOR inhibition uncouples HIV-1 latency reversal from cytokine-associated toxicity. J. Clin. Investig. 2017, 127, 651–656. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, F.Z.; Ouyang, Z.; Buzon, M.; Walker, B.D.; Lichterfeld, M.; Yu, X.G. Metabolic pathway activation distinguishes transcriptional signatures of CD8+ T cells from HIV-1 elite controllers. AIDS 2018, 32, 2669–2677. [Google Scholar] [CrossRef]
- Jin, S.; Liao, Q.; Chen, J.; Zhang, L.; He, Q.; Zhu, H.; Zhang, X.; Xu, J. TSC1 and DEPDC5 regulate HIV-1 latency through the mTOR signaling pathway. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Van Montfort, T.; van der Sluis, R.; Darcis, G.; Beaty, D.; Groen, K.; Pasternak, A.O.; Pollakis, G.; Vink, M.; Westerhout, E.M.; Hamdi, M.; et al. Dendritic cells potently purge latent HIV-1 beyond TCR-stimulation, activating the PI3K-Akt-mTOR pathway. EBioMedicine 2019, 42, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Reeves, D.B.; Duke, E.R.; Wagner, T.A.; Palmer, S.E.; Spivak, A.M.; Schiffer, J.T. A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation. Nat. Commun. 2018, 9, 4811. [Google Scholar] [CrossRef] [Green Version]
- Shiels, M.S.; Engels, E.A. Evolving epidemiology of HIV-associated malignancies. Curr. Opin. HIV AIDS 2017, 12, 6–11. [Google Scholar] [CrossRef]
- Tomlinson, C.C.; Damania, B. The K1 Protein of Kaposi’s Sarcoma-Associated Herpesvirus Activates the Akt Signaling Pathway. J. Virol. 2004, 78, 1918–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montaner, S. Akt/TSC/mTOR activation by the KSHV G protein-coupled receptor: Emerging insights into the molecular oncogenesis and treatment of Kaposi’s sarcoma. Cell Cycle 2007, 6, 438–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerr, D.A.; Busarla, S.V.P.; Gimbel, D.C.; Sohani, A.R.; Nazarian, R.M. mTOR, VEGF, PDGFR, and c-kit signaling pathway activation in Kaposi sarcoma. Hum. Pathol. 2017, 65, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.; Ganem, D. A Unique Herpesviral Transcriptional Program in KSHV-Infected Lymphatic Endothelial Cells Leads to mTORC1 Activation and Rapamycin Sensitivity. Cell Host Microbe 2013, 13, 429–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sin, S.-H.; Roy, D.; Wang, L.; Staudt, M.R.; Fakhari, F.D.; Patel, D.D.; Henry, D.; Harrington, W.J.; Damania, B.A.; Dittmer, D.P. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines in vivo by inhibiting autocrine signaling. Blood 2007, 109, 2165–2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Re, A.; Cattaneo, C.; Rossi, G. HIV and lymphoma: From epidemiology to clinical management. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019004. [Google Scholar] [CrossRef]
- Wang, X.; Duan, Z.; Yu, G.; Fan, M.; Scharff, M.D. Human Immunodeficiency Virus Tat Protein Aids V Region Somatic Hypermutation in Human B Cells. MBio 2018, 9, e02315–e02317. [Google Scholar] [CrossRef] [Green Version]
- El-Salem, M.; Raghunath, P.N.; Marzec, M.; Liu, X.; Kasprzycka, M.; Robertson, E.; Wasik, M.A. Activation of mTORC1 signaling pathway in AIDS-related lymphomas. Am. J. Pathol. 2009, 175, 817–824. [Google Scholar] [CrossRef] [Green Version]
- Sebestyén, A.; Sticz, T.B.; Márk, Á.; Hajdu, M.; Timár, B.; Nemes, K.; Nagy, N.; Váradi, Z.; Kopper, L. Activity and complexes of mTOR in diffuse large B-cell lymphomas—A tissue microarray study. Mod. Pathol. 2012, 25, 1623–1628. [Google Scholar] [CrossRef] [Green Version]
- Ágnes Márk, M.H.; Zsófia, V.; Sticz, T.B.; Nagy, N.; Csomor, J.; Berczi, L.; Varga, V.; Csóka, M.; Sebestyén, L.K.A. Characteristic mTOR activity in Hodgkin-lymphomas offers a potential therapeutic target in high risk disease—A combined tissue microarray, in vitro and in vivo study. BMC Cancer 2013, 13, 250. [Google Scholar]
- Browne, S.H.; Diaz-Perez, J.A.; Preziosi, M.; King, C.C.; Jones, G.A.; Jain, S.; Sun, X.; Reid, E.G.; Vandenberg, S.; Wang, H.Y. mTOR activity in AIDS-related diffuse large B-cell lymphoma. PLoS ONE 2017, 12, e0170771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Col, J.D.; Zancai, P.; Terrin, L.; Guidoboni, M.; Ponzoni, M.; Pavan, A.; Spina, M.; Bergamin, S.; Rizzo, S.; Tirelli, U.; et al. Distinct functional significance of Akt and mTOR constitutive activation in mantle cell lymphoma. Blood 2008, 111, 5142–5151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekihara, K.; Saitoh, K.; Han, L.; Ciurea, S.; Yamamoto, S.; Kikkawa, M.; Kazuno, S.; Taka, H.; Kaga, N.; Arai, H.; et al. Targeting mantle cell lymphoma metabolism and survival through simultaneous blockade of mTOR and nuclear transporter exportin-1. Oncotarget 2017, 8, 34552–34564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, G.; Herbrecht, R.; Romaguera, J.; Verhoef, G.; Crump, M.; Gisselbrecht, C.; Laurell, A.; Offner, F.; Strahs, A.; Berkenblit, A.; et al. Phase III Study to Evaluate Temsirolimus Compared With Investigator’s Choice Therapy for the Treatment of Relapsed or Refractory Mantle Cell Lymphoma. J. Clin. Oncol. 2009, 27, 3822–3829. [Google Scholar] [CrossRef] [PubMed]
- Witzig, T.E.; Reeder, C.B.; LaPlant, B.R.; Gupta, M.; Johnston, P.B.; Micallef, I.N.; Porrata, L.F.; Ansell, S.M.; Colgan, J.P.; Jacobsen, E.D.; et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia 2011, 25, 341–347. [Google Scholar] [CrossRef]
- Johnston, P.B.; LaPlant, B.; McPhail, E.; Habermann, T.M.; Inwards, D.J.; Micallef, I.N.; Colgan, J.P.; Nowakowski, G.S.; Ansell, S.M.; Witzig, T.E. Everolimus combined with R-CHOP-21 for new, untreated, diffuse large B-cell lymphoma (NCCTG 1085 [Alliance]): Safety and efficacy results of a phase 1 and feasibility trial. Lancet Haematol. 2016, 3, e309–e316. [Google Scholar] [CrossRef] [Green Version]
- Bennani, N.N.; LaPlant, B.R.; Ansell, S.M.; Habermann, T.M.; Inwards, D.J.; Micallef, I.N.; Johnston, P.B.; Porrata, L.F.; Colgan, J.P.; Markovic, S.N.; et al. Efficacy of the oral mTORC1 inhibitor everolimus in relapsed or refractory indolent lymphoma. Am. J. Hematol. 2017, 92, 448–453. [Google Scholar] [CrossRef] [Green Version]
- Johnston, P.B.; Pinter-Brown, L.C.; Warsi, G.; White, K.; Ramchandren, R. Phase 2 study of everolimus for relapsed or refractory classical Hodgkin lymphoma. Exp. Hematol. Oncol. 2018, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Faes, S.; Demartines, N.; Dormond, O. Resistance to mTORC1 Inhibitors in Cancer Therapy: From Kinase Mutations to Intratumoral Heterogeneity of Kinase Activity. Oxid. Med. Cell. Longev. 2017, 2017, 1726078. [Google Scholar] [CrossRef] [Green Version]
- Petrich, A.M.; Leshchenko, V.; Kuo, P.-Y.; Xia, B.; Thirukonda, V.K.; Ulahannan, N.; Gordon, S.; Fazzari, M.J.; Ye, B.H.; Sparano, J.A.; et al. Akt Inhibitors MK-2206 and Nelfinavir Overcome mTOR Inhibitor Resistance in Diffuse Large B-cell Lymphoma. Clin. Cancer Res. 2012, 18, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Heredia, A.; Gilliam, B.; Latinovic, O.; Le, N.; Bamba, D.; DeVico, A.; Melikyan, G.B.; Gallo, R.C.; Redfield, R.R. Rapamycin reduces CCR5 density levels on CD4 T cells, and this effect results in potentiation of enfuvirtide (T-20) against R5 strains of human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 2007, 51, 2489–2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heredia, A.; Latinovic, O.; Gallo, R.C.; Melikyan, G.; Reitz, M.; Le, N.; Redfield, R.R. Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc. Natl. Acad. Sci. USA 2008, 105, 20476–20481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Konkimalla, S.; Yadav, A.; Sataranatarajan, K.; Kasinath, B.S.; Chander, P.N.; Singhal, P.C. HIV-Associated Nephropathy. Am. J. Pathol. 2010, 177, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Husain, M.; Yadav, A.; Kasinath, B.S.; Malhotra, A.; Singhal, P.C. HIV-1 Promotes Renal Tubular Epithelial Cell Protein Synthesis: Role of mTOR Pathway. PLoS ONE 2012, 7, e30071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.; Rai, P.; Plagov, A.; Lan, X.; Mathieson, P.W.; Saleem, M.A.; Husain, M.; Malhotra, A.; Singhal, P.C. Rapamycin-induced modulation of miRNA expression is associated with amelioration of HIV-associated nephropathy (HIVAN). Exp. Cell Res. 2013, 319, 2073–2080. [Google Scholar] [CrossRef] [Green Version]
- Rai, P.; Plagov, A.; Lan, X.; Chandel, N.; Singh, T.; Lederman, R.; Ayasolla, K.R.; Mathieson, P.W.; Saleem, M.A.; Husain, M.; et al. mTOR plays a critical role in p53-induced oxidative kidney cell injury in HIVAN. Am. J. Physiol. Physiol. 2013, 305, F343–F354. [Google Scholar] [CrossRef] [Green Version]
- Rai, P.; Lederman, R.; Haque, S.; Rehman, S.; Kumar, V.; Sataranatrajan, K.; Malhotra, A.; Kasinath, B.S.; Singhal, P.C. Renin angiotensin system modulates mTOR pathway through AT2R in HIVAN. Exp. Mol. Pathol. 2014, 96, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Department of Health and Human Services. Panel on Antiretroviral Guidelines for Adults and Adolescents.Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Available online: https://aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf (accessed on 10 April 2020).
- Shim, J.S.; Rao, R.; Beebe, K.; Neckers, L.; Han, I.; Nahta, R.; Liu, J.O. Selective Inhibition of HER2-Positive Breast Cancer Cells by the HIV Protease Inhibitor Nelfinavir. JNCI J. Natl. Cancer Inst. 2012, 104, 1576–1590. [Google Scholar] [CrossRef] [Green Version]
- Srirangam, A.; Mitra, R.; Wang, M.; Gorski, J.C.; Badve, S.; Baldridge, L.; Hamilton, J.; Kishimoto, H.; Hawes, J.; Li, L.; et al. Effects of HIV protease inhibitor ritonavir on Akt-regulated cell proliferation in breast cancer. Clin. Cancer Res. 2006, 12, 1883–1896. [Google Scholar] [CrossRef] [Green Version]
- Goda, J.; Pachpor, T.; Basu, T.; Chopra, S.; Gota, V. Targeting the AKT pathway: Repositioning HIV protease inhibitors as radiosensitizers. Indian J. Med. Res. 2016, 143, 145. [Google Scholar] [CrossRef] [Green Version]
- Aldinucci, D.; Casagrande, N. Inhibition of the CCL5/CCR5 axis against the progression of gastric cancer. Int. J. Mol. Sci. 2018, 19, 1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, D.; Rahbar, R.; Fish, E.N. CCL5 activation of CCR5 regulates cell metabolism to enhance proliferation of breast cancer cells. Open Biol. 2016, 6, 160122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.J. Off-target effects of drugs that disrupt human mitochondrial DNA maintenance. Front. Mol. Biosci. 2017, 4, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margolis, A.M.; Heverling, H.; Pham, P.A.; Stolbach, A. A Review of the Toxicity of HIV Medications. J. Med. Toxicol. 2014, 10, 26–39. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Stankov, M.V.; Hegermann, J.; Budida, R.; Panayotova-Dimitrova, D.; Schmidt, R.E.; Behrensa, G.M.N. Zidovudine-mediated autophagy inhibition enhances mitochondrial toxicity in muscle cells. Antimicrob. Agents Chemother. 2019, 63, e01443-18. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Wu, L.Y. Influence of phosphatidylinositol-3-kinase/protein kinase b-mammalian target of rapamycin signaling pathway on the neuropathic pain complicated by nucleoside reverse transcriptase inhibitors for the treatment of HIV infection. Chin. Med. J. (Engl.) 2018, 131, 1849–1856. [Google Scholar] [CrossRef]
- Hardy, W.D. Fundamentals of HIV Medicine 2019; Oxford University Press: Oxford, UK, 2019; ISBN 9780190942519. [Google Scholar]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbay, B.; Shmakova, A.; Vassetzky, Y.; Dokudovskaya, S. Modulation of mTORC1 Signaling Pathway by HIV-1. Cells 2020, 9, 1090. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9051090
Akbay B, Shmakova A, Vassetzky Y, Dokudovskaya S. Modulation of mTORC1 Signaling Pathway by HIV-1. Cells. 2020; 9(5):1090. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9051090
Chicago/Turabian StyleAkbay, Burkitkan, Anna Shmakova, Yegor Vassetzky, and Svetlana Dokudovskaya. 2020. "Modulation of mTORC1 Signaling Pathway by HIV-1" Cells 9, no. 5: 1090. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9051090