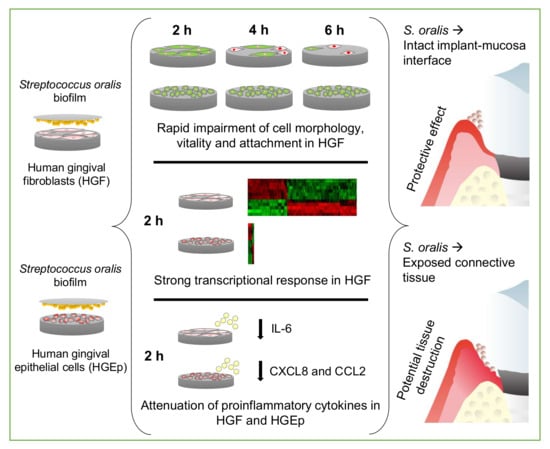
In Vitro Effects of Streptococcus oralis Biofilm on Peri-Implant Soft Tissue Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Formation of Streptococcus oralis Biofilm
2.2. Human Tissue Cell Culture and Titanium Colonization
2.3. Co-Culture of Tissue Cells Grown on Titanium with Streptococcus oralis Biofilm
2.4. Qualitative LIVE/DEAD Staining and Quantitative Analysis of Cell Attachment of Gingival Tissue Cells after Biofilm Challenge
2.5. RNA Extraction of Tissue Cells
2.6. Array Hybridization and Data Analysis
2.7. Cytokine Detection in Co-Culture Supernatants
2.8. Statistical Analysis
3. Results
3.1. Cell Morphology, Vitality, and Attachment of HGF were Rapidly Impaired After Challenge with the S. oralis Biofilm, whereas HGEp Remained Unaffected
3.2. Strong Transcriptional Response to Commensal S. oralis biofilm in HGF Compared to HGEp
3.3. Attenuation of Proinflammatory Cytokines in HGF and HGEp after Their Co-Culture with S. oralis Biofilm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Belibasakis, G.N. Microbiological and immuno-pathological aspects of peri-implant diseases. Arch. Oral Biol. 2014, 59, 66–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irshad, M.; Scheres, N.; Anssari Moin, D.; Crielaard, W.; Loos, B.G.; Wismeijer, D.; Laine, M.L. Cytokine and matrix metalloproteinase expression in fibroblasts from peri-implantitis lesions in response to viable Porphyromonas gingivalis. J. Periodontal Res. 2013, 48, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Charalampakis, G.; Belibasakis, G.N. Microbiome of peri-implant infections: Lessons from conventional, molecular and metagenomic analyses. Virulence 2015, 6, 183–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, M.G.; Lindhe, J. Peri-implant health. J. Clin. Periodontol. 2018, 45, S230–S236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belibasakis, G.N.; Charalampakis, G.; Bostanci, N.; Stadlinger, B. Peri-implant infections of oral biofilm etiology. Adv. Exp. Med. Biol. 2015, 830, 69–84. [Google Scholar] [PubMed]
- Berglundh, T.; Armitage, G.; Araujo, M.G.; Avila-Ortiz, G.; Blanco, J.; Camargo, P.M.; Chen, S.; Cochran, D.; Derks, J.; Figuero, E.; et al. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89, S313–S328. [Google Scholar] [CrossRef] [PubMed]
- Kinikoglu, B.; Damour, O.; Hasirci, V. Tissue engineering of oral mucosa: A shared concept with skin. J. Artif. Organs 2015, 18, 8–19. [Google Scholar] [CrossRef]
- Dale, B.A. Fascination with Epithelia: Architecture, Proteins, and Functions. J. Dent. Res. 2003, 82, 866–869. [Google Scholar] [CrossRef]
- Abiko, Y.; Hiratsuka, K.; Kiyama-Kishikawa, M.; Tsushima, K.; Ohta, M.; Sasahara, H. Profiling of differentially expressed genes in human gingival epithelial cells and fibroblasts by DNA microarray. J. Oral Sci. 2004, 46, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Belibasakis, G.N.; Bao, K.; Bostanci, N. Transcriptional profiling of human gingival fibroblasts in response to multi-species in vitro subgingival biofilms. Mol. Oral Microbiol. 2014, 29, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Palaiologou, A.A.; Yukna, R.A.; Moses, R.; Lallier, T.E. Gingival, Dermal, and Periodontal Ligament Fibroblasts Express Different Extracellular Matrix Receptors. J. Periodontol. 2001, 72, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, J.; Menzel, N.; Dommisch, H.; Winter, J.; Jepsen, S.; Mutters, R. The stage of native biofilm formation determines the gene expression of human beta-defensin-2, psoriasin, ribonuclease 7 and inflammatory mediators: A novel approach for stimulation of keratinocytes with in situ formed biofilms. Oral Microbiol. Immunol. 2008, 23, 21–28. [Google Scholar] [CrossRef]
- Guggenheim, B.; Gmur, R.; Galicia, J.C.; Stathopoulou, P.G.; Benakanakere, M.R.; Meier, A.; Thurnheer, T.; Kinane, D.F. In vitro modeling of host-parasite interactions: The ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol. 2009, 9, 280–2180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millhouse, E.; Jose, A.; Sherry, L.; Lappin, D.F.; Patel, N.; Middleton, A.M.; Pratten, J.; Culshaw, S.; Ramage, G. Development of an in vitro periodontal biofilm model for assessing antimicrobial and host modulatory effects of bioactive molecules. BMC Oral Health 2014, 14, 80–6831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramage, G.; Lappin, D.F.; Millhouse, E.; Malcolm, J.; Jose, A.; Yang, J.; Bradshaw, D.J.; Pratten, J.R.; Culshaw, S. The epithelial cell response to health and disease associated oral biofilm models. J. Periodont. Res. 2017, 52, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, Y.; Mans, J.J.; Mao, S.; Lopez, M.C.; Baker, H.V.; Handfield, M.; Lamont, R.J. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect. Immun. 2007, 75, 2540–2547. [Google Scholar] [CrossRef] [Green Version]
- Ebersole, J.L.; Peyyala, R.; Gonzalez, O.A. Biofilm-induced profiles of immune response gene expression by oral epithelial cells. Mol. Oral Microbiol. 2019, 34. [Google Scholar] [CrossRef] [Green Version]
- Furst, M.M.; Salvi, G.E.; Lang, N.P.; Persson, G.R. Bacterial colonization immediately after installation on oral titanium implants. Clin. Oral Implants Res. 2007, 18, 501–508. [Google Scholar] [CrossRef]
- Gomez-Florit, M.; Ramis, J.M.; Xing, R.; Taxt-Lamolle, S.; Haugen, H.J.; Lyngstadaas, S.P.; Monjo, M. Differential response of human gingival fibroblasts to titanium- and titanium-zirconium-modified surfaces. J. Periodontal Res. 2014, 49, 425–436. [Google Scholar] [CrossRef]
- Diaz, P.I.; Chalmers, N.I.; Rickard, A.H.; Kong, C.; Milburn, C.L.; Palmer, R.J.; Kolenbrander, P.E. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 2006, 72, 2837–2848. [Google Scholar] [CrossRef] [Green Version]
- Diaz, P.I.; Xie, Z.; Sobue, T.; Thompson, A.; Biyikoglu, B.; Ricker, A.; Ikonomou, L.; Dongari-Bagtzoglou, A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 2012, 80, 620–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Socransky, S.S.; Haffajee, A.D. Dental biofilms: Difficult therapeutic targets. Periodontology 2000, 28, 12–55. [Google Scholar] [CrossRef] [PubMed]
- Carrouel, F.; Viennot, S.; Santamaria, J.; Veber, P.; Bourgeois, D. Quantitative Molecular Detection of 19 Major Pathogens in the Interdental Biofilm of Periodontally Healthy Young Adults. Front. Microbiol. 2016, 7, 840. [Google Scholar] [CrossRef] [PubMed]
- Boutin, S.; Hagenfeld, D.; Zimmermann, H.; El Sayed, N.; Hopker, T.; Greiser, H.K.; Becher, H.; Kim, T.S.; Dalpke, A.H. Clustering of Subgingival Microbiota Reveals Microbial Disease Ecotypes Associated with Clinical Stages of Periodontitis in a Cross-Sectional Study. Front. Microbiol. 2017, 8, 340. [Google Scholar] [CrossRef] [Green Version]
- Douglas, C.W.; Heath, J.; Hampton, K.K.; Preston, F.E. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 1993, 39, 179–182. [Google Scholar] [CrossRef] [Green Version]
- Beighton, D.; Carr, A.D.; Oppenheim, B.A. Identification of viridans streptococci associated with bacteraemia in neutropenic cancer patients. J. Med. Microbiol. 1994, 40, 202–204. [Google Scholar] [CrossRef]
- Ahmed, R.; Hassall, T.; Morland, B.; Gray, J. Viridans streptococcus bacteremia in children on chemotherapy for cancer: An underestimated problem. Pediatr. Hematol. Oncol. 2003, 20, 439–444. [Google Scholar] [CrossRef]
- Dyson, C.; Barnes, R.A.; Harrison, G.A.J. Infective endocarditis: An epidemiological review of 128 episodes. J. Infect. 1999, 38, 87–93. [Google Scholar] [CrossRef]
- Koren, O.; Spor, A.; Felin, J.; Fak, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4592–4598. [Google Scholar] [CrossRef] [Green Version]
- Lehtiniemi, J.; Karhunen, P.J.; Goebeler, S.; Nikkari, S.; Nikkari, S.T. Identification of different bacterial DNAs in human coronary arteries. Eur. J. Clin. Investig. 2005, 35, 13–16. [Google Scholar] [CrossRef]
- Nagata, E.; de Toledo, A.; Oho, T. Invasion of human aortic endothelial cells by oral viridans group streptococci and induction of inflammatory cytokine production. Mol. Oral Microbiol. 2011, 26, 78–88. [Google Scholar] [CrossRef] [PubMed]
- De Toledo, A.; Nagata, E.; Yoshida, Y.; Oho, T. Streptococcus oralis coaggregation receptor polysaccharides induce inflammatory responses in human aortic endothelial cells. Mol. Oral Microbiol. 2012, 27, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Ammann, T.W.; Belibasakis, G.N.; Thurnheer, T. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS ONE 2013, 8, e83090. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Andersen, R.N.; Blehert, D.S.; Egland, P.G.; Foster, J.S.; Palmer, R.J. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 486–505. [Google Scholar] [CrossRef] [Green Version]
- Whitmore, S.E.; Lamont, R.J. The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 2011, 81, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Sobue, T.; Thompson, A.; Xie, Z.; Poon, K.; Ricker, A.; Cervantes, J.; Diaz, P.I.; Dongari-Bagtzoglou, A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014, 16, 214–231. [Google Scholar] [CrossRef]
- Xu, H.; Sobue, T.; Bertolini, M.; Thompson, A.; Dongari-Bagtzoglou, A. Streptococcus oralis and Candida albicans Synergistically Activate mu-Calpain to Degrade E-cadherin From Oral Epithelial Junctions. J. Infect. Dis. 2016, 214, 925–934. [Google Scholar] [CrossRef] [Green Version]
- Koo, H.; Andes, D.R.; Krysan, D.J. Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 2018, 14, e1007342. [Google Scholar] [CrossRef] [Green Version]
- Thurnheer, T.; Belibasakis, G.N. Streptococcus oralis maintains homeostasis in oral biofilms by antagonizing the cariogenic pathogen Streptococcus mutans. Mol. Oral Microbiol. 2018, 33, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Bidossi, A.; De Grandi, R.; Toscano, M.; Bottagisio, M.; De Vecchi, E.; Gelardi, M.; Drago, L. Probiotics Streptococcus salivarius 24SMB and Streptococcus oralis 89a interfere with biofilm formation of pathogens of the upper respiratory tract. BMC Infect. Dis. 2018, 18, 653. [Google Scholar] [CrossRef]
- Cosseau, C.; Devine, D.A.; Dullaghan, E.; Gardy, J.L.; Chikatamarla, A.; Gellatly, S.; Yu, L.L.; Pistolic, J.; Falsafi, R.; Tagg, J.; et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 2008, 76, 4163–4175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Chen, R.; Rudney, J.D. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J. Periodontal Res. 2008, 43, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, R.; Rudney, J.D. Streptococcus cristatus modulates the Fusobacterium nucleatum-induced epithelial interleukin-8 response through the nuclear factor-kappa B pathway. J. Periodontal Res. 2011, 46, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, J.; Wang, Q.; Fitzsimonds, Z.R.; Miller, D.P.; Sztukowska, M.N.; Jung, Y.J.; Hayashi, M.; Whiteley, M.; Lamont, R.J. Streptococcus gordonii programs epithelial cells to resist ZEB2 induction by Porphyromonas gingivalis. Proc. Natl. Acad. Sci. USA 2019, 116, 8544–8553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darveau, R.P. Periodontitis: A polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010, 8, 481–490. [Google Scholar] [CrossRef]
- Devine, D.A.; Marsh, P.D.; Meade, J. Modulation of host responses by oral commensal bacteria. J. Oral Microbiol. 2015, 7. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G.; Brouwer, C. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef]
- Berglundh, T.; Lindhe, J.; Ericsson, I.; Marinello, C.P.; Liljenberg, B.; Thomsen, P. The soft tissue barrier at implants and teeth. Clin. Oral Implants Res. 1991, 2, 81–90. [Google Scholar] [CrossRef]
- Groeger, S.E.; Meyle, J. Epithelial barrier and oral bacterial infection. Periodontology 2000, 69, 46–67. [Google Scholar] [CrossRef]
- Schincaglia, G.P.; Hong, B.Y.; Rosania, A.; Barasz, J.; Thompson, A.; Sobue, T.; Panagakos, F.; Burleson, J.A.; Dongari-Bagtzoglou, A.; Diaz, P.I. Clinical, Immune, and Microbiome Traits of Gingivitis and Peri-implant Mucositis. J. Dent. Res. 2017, 96, 47–55. [Google Scholar] [CrossRef]
- Sukhithasri, V.; Nisha, N.; Biswas, L.; Anil Kumar, V.; Biswas, R. Innate immune recognition of microbial cell wall components and microbial strategies to evade such recognitions. Microbiol. Res. 2013, 168, 396–406. [Google Scholar] [CrossRef]
- Uehara, A.; Sugawara, Y.; Kurata, S.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Satta, Y.; Sasano, T.; Sugawara, S.; Takada, H. Chemically synthesized pathogen-associated molecular patterns increase the expression of peptidoglycan recognition proteins via toll-like receptors, NOD1 and NOD2 in human oral epithelial cells. Cell Microbiol. 2005, 7, 675–686. [Google Scholar] [CrossRef]
- Uehara, A.; Takada, H. Functional TLRs and NODs in human gingival fibroblasts. J. Dent. Res. 2007, 86, 249–254. [Google Scholar] [CrossRef]
- Sugawara, Y.; Uehara, A.; Fujimoto, Y.; Kusumoto, S.; Fukase, K.; Shibata, K.; Sugawara, S.; Sasano, T.; Takada, H. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J. Dent. Res. 2006, 85, 524–529. [Google Scholar] [CrossRef]
- Mans, J.J.; von Lackum, K.; Dorsey, C.; Willis, S.; Wallet, S.M.; Baker, H.V.; Lamont, R.J.; Handfield, M. The degree of microbiome complexity influences the epithelial response to infection. BMC Genom. 2009, 10, 380–2164. [Google Scholar] [CrossRef] [Green Version]
- Handfield, M.; Mans, J.J.; Zheng, G.; Lopez, M.C.; Mao, S.; Progulske-Fox, A.; Narasimhan, G.; Baker, H.V.; Lamont, R.J. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005, 7, 811–823. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Anderson, P. Post-transcriptional control of cytokine production. Nat. Immunol. 2008, 9, 353–359. [Google Scholar] [CrossRef]
- Stow, J.L.; Murray, R.Z. Intracellular trafficking and secretion of inflammatory cytokines. Cytokine Growth Factor Rev. 2013, 24, 227–239. [Google Scholar] [CrossRef]
- Basso, F.G.; Pansani, T.N.; Turrioni, A.P.; Soares, D.G.; de Souza Costa, C.A.; Hebling, J. Tumor Necrosis Factor-α and Interleukin (IL)-1β, IL-6, and IL-8 Impair In Vitro Migration and Induce Apoptosis of Gingival Fibroblasts and Epithelial Cells, Delaying Wound Healing. J. Periodontol. 2016, 87, 990–996. [Google Scholar] [CrossRef]
- Naruishi, K.; Nagata, T. Biological effects of interleukin-6 on Gingival Fibroblasts: Cytokine regulation in periodontitis. J. Cell. Physiol. 2018, 233, 6393–6400. [Google Scholar] [CrossRef]
- Suchett-Kaye, G.; Morrier, J.J.; Barsotti, O. Interactions between non-immune host cells and the immune system during periodontal disease: Role of the gingival keratinocyte. Crit. Rev. Oral Biol. Med. 1998, 9, 292–305. [Google Scholar] [CrossRef]
- Pan, W.; Wang, Q.; Chen, Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Twetman, S.; Derawi, B.; Keller, M.; Ekstrand, K.; Yucel-Lindberg, T.; Stecksen-Blicks, C. Short-term effect of chewing gums containing probiotic Lactobacillus reuteri on the levels of inflammatory mediators in gingival crevicular fluid. Acta Odontol. Scand. 2009, 67, 19–24. [Google Scholar] [CrossRef]
- La Mantia, I.; Varricchio, A.; Ciprandi, G. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for preventing recurrent acute otitis media in children: A real-life clinical experience. Int. J. Gen. Med. 2017, 10, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Andaloro, C.; Santagati, M.; Stefani, S.; La Mantia, I. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a oral spray for children with recurrent streptococcal pharyngotonsillitis: A randomized placebo-controlled clinical study. Eur. Arch. Otorhinolaryngol. 2019, 276, 879–887. [Google Scholar] [CrossRef]
- Patini, R.; Cattani, P.; Marchetti, S.; Isola, G.; Quaranta, G.; Gallenzi, P. Evaluation of Predation Capability of Periodontopathogens Bacteria by Bdellovibrio Bacteriovorus HD100. An in Vitro Study. Materials (Basel) 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Basso, F.G.; Soares, D.G.; Pansani, T.N.; Turrioni, A.P.S.; Scheffel, D.L.; de Souza Costa, C.A.; Hebling, J. Effect of LPS treatment on the viability and chemokine synthesis by epithelial cells and gingival fibroblasts. Arch. Oral Biol. 2015, 60, 1117–1121. [Google Scholar] [CrossRef]
- Uehara, A.; Fujimoto, Y.; Fukase, K.; Takada, H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol. Immunol. 2007, 44, 3100–3111. [Google Scholar] [CrossRef]
- Homer, K.A.; Whiley, R.A.; Beighton, D. Proteolytic activity of oral streptococci. FEMS Microbiol. Lett. 1990, 55, 257–260. [Google Scholar] [CrossRef]
- Beighton, D.; Homer, K.A.; Kelley, S. The Production of Protease Activities by Streptococcus oralis Strains Isolated from Endocarditis. Microb. Ecol. Health Dis. 1995, 8, 213–218. [Google Scholar] [CrossRef] [Green Version]
- Rafay, A.M.; Homer, K.A.; Beighton, D. Effect of mucin and glucose on proteolytic and glycosidic activities of Streptococcus oralis. J. Med. Microbiol. 1996, 44, 409–417. [Google Scholar] [CrossRef]
- Stavroullakis, A.; Brito, C.; Chen, H.Y.; Bajenova, E.; Prakki, A.; Nogueira-Filho, G. Dental implant surface treatments may modulate cytokine secretion in Porphyromonas gingivalis-stimulated human gingival fibroblasts: A comparative study. J. Biomed. Mater. Res. A 2015, 103, 1131–1140. [Google Scholar] [CrossRef]





| Human Gingival Fibroblasts | Human Gingival Epithelial Cells | |||||
|---|---|---|---|---|---|---|
| Pathway | % | p | Genes | % | p | Genes |
| Chemokine signaling pathway | 5.32 | 0.0421 | CXCL1L, CCL2L, NCF1RS, CXCL3L, CXCL2L | 0.25 | 0.0068 | CXCL1L, CXCL2L, CXCL8L |
| Cytokine-cytokine receptor interaction | 5.32 | 0.0791 | CSF2L, IL6L, CCL2L, IL12AL, TNFSF9L | 0.25 | 0.0103 | IL6L, TNFL, CXCL8L |
| FoxO signaling pathway | 7.45 | 0.0003 | SGK1TI, IL6L, PLK3RS, PLK2RS, GADD45GRS, GADD45BRS, KLF2RS | |||
| Jak-STAT signaling pathway | 5.32 | 0.0190 | CSF2L, IL6L, SOCS3TI, IL12AL, MYCTF | |||
| MAPK signaling pathway | 10.64 | 0.0001 | DUSP5TI, FGF5L, FOSTF, DUSP2TI, DUSP1TI, JUNTF, GADD45GTA, GADD45BTA, MYCTF, NGFL | |||
| NF-kappa B signaling pathway | 0.17 | 0.0614 | TNFL, CXCL8RS | |||
| NOD-like receptor signaling pathway | 0.25 | 0.0006 | IL6RS, TNFRS, CXCL8RS | |||
| p53 signaling pathway | 3.19 | 0.0814 | GADD45GTI, RS, PMAIP1TA, GADD45BTI, RS | |||
| PI3K-Akt signaling pathway | 6.38 | 0.0959 | FGF5L, SGK1TA, IL6L, MYCTF, EPHA2RC, NGFL | |||
| TNF signaling pathway | 11.70 | 0.0000 | CXCL1RS, CSF2RS, FOSTF, RS, IL6RS, CCL2RS, SOCS3RS, CXCL3RS, JUNTF, RS, EDN1RS, CXCL2RS, JUNBTF,RS | 0.34 | 0.0000 | CXCL1RS, IL6L, TNFL, CXCL2RS |
| Toll-like receptor signaling pathway | 4.26 | 0.0389 | FOSTF, IL6RS, JUNTF, IL12ARS | 0.25 | 0.0023 | IL6RS, TNFRS, CXCL8RS |
| Wnt signaling pathway | 4.26 | 0.0739 | DKK1TI, JUNTF, BAMBITI, MYCTF | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingendoh-Tsakmakidis, A.; Eberhard, J.; Falk, C.S.; Stiesch, M.; Winkel, A. In Vitro Effects of Streptococcus oralis Biofilm on Peri-Implant Soft Tissue Cells. Cells 2020, 9, 1226. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9051226
Ingendoh-Tsakmakidis A, Eberhard J, Falk CS, Stiesch M, Winkel A. In Vitro Effects of Streptococcus oralis Biofilm on Peri-Implant Soft Tissue Cells. Cells. 2020; 9(5):1226. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9051226
Chicago/Turabian StyleIngendoh-Tsakmakidis, Alexandra, Jörg Eberhard, Christine S. Falk, Meike Stiesch, and Andreas Winkel. 2020. "In Vitro Effects of Streptococcus oralis Biofilm on Peri-Implant Soft Tissue Cells" Cells 9, no. 5: 1226. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9051226