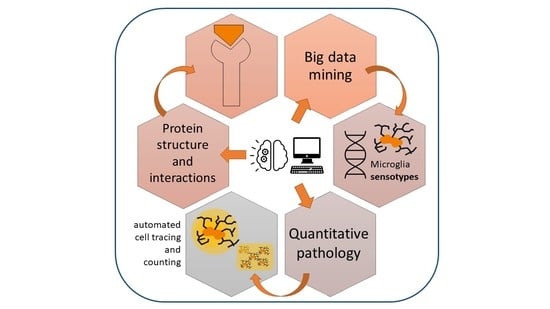
Brain Immunoinformatics: A Symmetrical Link between Informatics, Wet Lab and the Clinic
Abstract
:1. Introduction
2. Literature Search and Field Definition
- Original publications and methods that contributed to breakthrough observations in the field (result-oriented selection approach), such as the TYROBP role in Alzheimer’s disease;
- Well-renowned public databases that set a new cornerstone for data science (data-driven approach);
- Outstanding longitudinal works that reshaped the doctrines of neuroimmunology in the last decade, such as the innovative classification of microglial cell activation and the development of image reconstruction tools (longitudinal approach).
3. Big Data-Omics in Neuroimmunoinformatics
3.1. Genome-Wide Studies
3.2. Single-Cell Studies
3.3. Expression Quantitative Trait Loci
3.4. Epigenetics, ChIP-Seq, and ATAC-Seq
4. Microglial “Next-Generation” Classification: From “Resting” to “Active”, to “Polarized”, to “De Novo Classified”
Examples of Microglial Sensotypes
- Homeostatic/resting sensotype or “sensing” MG: is the sensotype that differentiates MG from MΦ in the healthy brain, based on gene expression. Such differentiation is morphologically not possible. Transcripts such as Hexb, Tmem119, Siglech, P2ry12, and Olfm13 are unique to mice MG, distinguishing them from brain MΦ [4,28,55,56]. Cx3cr1 is a gene with strong MG expression, suggested as a signature gene for MG, despite controversies [57]. Micro-RNAs (miRNA) such as miRNA99a, miRNA125b-5p, and miRNA342-3p are expressed only in MG and not in other myeloid cells in murine models [57], whereas the fingerprint of human versus murine MG is still a challenging research terrain [58]. Evidence converges towards Tyrobp as an important sensing factor [56], PU.1 as the most decisive transcription factor, and TGFβ as the most robust lineage upstream stimulator of human MG [4,51].
- Induced human MG (iMG): informatics has supported stem-cell technology and provided tools to characterize iMG in vitro, either in an austere environment or in co-culture with h-iPSC organoids. Besides co-culture induced changes such as SIGLECH upregulation and Tmem119/Tyrobp downregulation, scRNA-seq technology allowed for spotting iMG sensome differences between ventral and dorsal h-iPSC organoids, with significant overexpression of inflammatory genes such as TNFa, IL6, and TREM2 in the ventral organoid iMG [62].
- Xenografted human MG (xMG) in mouse brain chimeras; Xu et al. applied scRNA-seq to sensotype xenografted human brain microglia in MG-depleted mouse brain chimeras [63]. The xMG sensome was evidenced to retain microglial (TMEM119, P2Ry12, SALL1, and OLFML3) and hominid (SPP1, A2M, and C3) traits as xenograft [58].
5. Machine Learning for Prediction of Protein, Cellular, and Network Interactions
5.1. Protein-Epitope Affinity Prediction, Cytokine-Receptor Interactions, and Epitope Prediction
5.2. Cell-Cell Interactions and Multiscale Network Modeling
5.3. Probabilistic and Causal Gene Regulatory Networks
6. Machine Learning in Neuropathology and Immunoprofiling
6.1. Microglial Segmentation and Counting
6.2. Cell Arborization Analysis
6.3. Automated Cell Arbor Tracing
6.4. End-to-End Solutions for Microglia Characterization: Interfaces for Implementation by Biologists and Non-Computer Scientists
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hammerbacher, J.; Snyder, A. Informatics for cancer immunotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, xii56–xii73. [Google Scholar] [CrossRef]
- Jabbari, P.; Rezaei, N. Artificial intelligence and immunotherapy. Expert Rev. Clin. Immunol. 2019, 15, 689–691. [Google Scholar] [CrossRef]
- Richards, B.A.; Lillicrap, T.P.; Beaudoin, P.; Bengio, Y.; Bogacz, R.; Christensen, A.; Clopath, C.; Costa, R.P.; de Berker, A.; Ganguli, S.; et al. A deep learning framework for neuroscience. Nat. Neurosci. 2019, 22, 1761–1770. [Google Scholar] [CrossRef]
- Yeh, H.; Ikezu, T. Transcriptional and Epigenetic Regulation of Microglia in Health and Disease. Trends Mol. Med. 2019, 25, 96–111. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Walker, J.R.; Spies, N.C.; Ainscough, B.J.; Griffith, O.L. Informatics for RNA Sequencing: A Web Resource for Analysis on the Cloud. PLoS Comput. Biol. 2015, 11, e1004393. [Google Scholar] [CrossRef] [Green Version]
- Kidd, B.A.; Peters, L.A.; Schadt, E.E.; Dudley, J.T. Unifying immunology with informatics and multiscale biology. Nat. Immunol. 2014, 15, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Neu, K.E.; Tang, Q.; Wilson, P.C.; Khan, A.A. Single-Cell Genomics: Approaches and Utility in Immunology. Trends Immunol. 2017, 38, 140–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wes, P.D.; Holtman, I.R.; Boddeke, E.W.G.M.; Möller, T.; Eggen, B.J.L. Next generation transcriptomics and genomics elucidate biological complexity of microglia in health and disease. Glia 2016, 64, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Yuan, Y.; Parmigiani, G.; Suraokar, M.B.; Diao, L.; Wistuba, I.I.; Wang, W. DeMix: Deconvolution for mixed cancer transcriptomes using raw measured data. Bioinforma. Oxf. Engl. 2013, 29, 1865–1871. [Google Scholar] [CrossRef]
- Shen-Orr, S.S.; Tibshirani, R.; Khatri, P.; Bodian, D.L.; Staedtler, F.; Perry, N.M.; Hastie, T.; Sarwal, M.M.; Davis, M.M.; Butte, A.J. Cell type-specific gene expression differences in complex tissues. Nat. Methods 2010, 7, 287–289. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Heng, T.S.P.; Painter, M.W. Immunological Genome Project Consortium, The Immunological Genome Project: Networks of gene expression in immune cells. Nat. Immunol. 2008, 9, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [Green Version]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef] [PubMed]
- Elkjaer, M.L.; Frisch, T.; Reynolds, R.; Kacprowski, T.; Burton, M.; Kruse, T.A.; Thomassen, M.; Baumbach, J.; Illes, Z. Molecular signature of different lesion types in the brain white matter of patients with progressive multiple sclerosis. Acta Neuropathol. Commun. 2019, 7, 205. [Google Scholar] [CrossRef] [Green Version]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, A. Genome-Wide Association Studies. Methods Mol. Biol. Clifton NJ 2018, 1793, 37–49. [Google Scholar] [CrossRef]
- Flanagan, J.M. Epigenome-wide association studies (EWAS): Past, present, and future. Methods Mol. Biol. Clifton NJ 2015, 1238, 51–63. [Google Scholar] [CrossRef]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.I.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 Loci Associated with Bipolar Disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef]
- Ball, R.D. Statistical analysis of genomic data. Methods Mol. Biol. Clifton NJ 2013, 1019, 171–192. [Google Scholar] [CrossRef]
- Hayes, B. Overview of Statistical Methods for Genome-Wide Association Studies (GWAS). Methods Mol. Biol. Clifton NJ 2013, 1019, 149–169. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, L.; Liu, D.; Hu, S.; Song, X.; Li, J.; Lv, H.; Duan, L.; Zhang, M.; Jiang, Q.; et al. EWAS: Epigenome-wide association study software 2.0. Bioinforma. Oxf. Engl. 2018, 34, 2657–2658. [Google Scholar] [CrossRef]
- Arloth, J.; Eraslan, G.; Andlauer, T.F.M.; Martins, J.; Iurato, S.; Kühnel, B.; Waldenberger, M.; Frank, J.; Gold, R.; Hemmer, B.; et al. DeepWAS: Multivariate genotype-phenotype associations by directly integrating regulatory information using deep learning. PLoS Comput. Biol. 2020, 16, e1007616. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Yao, X.; Hu, Y.; Zhao, H. GenoWAP: GWAS signal prioritization through integrated analysis of genomic functional annotation. Bioinformatics 2016, 32, 542–548. [Google Scholar] [CrossRef]
- Hoffjan, S.; Okur, A.; Epplen, J.T.; Wieczorek, S.; Chan, A.; Akkad, D.A. Association of TNFAIP3 and TNFRSF1A variation with multiple sclerosis in a German case-control cohort. Int. J. Immunogenet. 2015, 42, 106–110. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinforma. Oxf. Engl. 2012, 28, 1353–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Development Core Team. A language and environment for statistical computing. In R Foundation for Statistical Computing; Vienna, Austria, 2021; Available online: http://www.R-project.org (accessed on 1 July 2021).
- Liu, Y.; Ye, Y.; Gong, J.; Han, L. Expression Quantitative Trait Loci (eQTL) Analysis in Cancer. Methods Mol. Biol. Clifton NJ 2020, 2082, 189–199. [Google Scholar] [CrossRef]
- Mirza, N.; Appleton, R.; Burn, S.; du Plessis, D.; Duncan, R.; Farah, J.O.; Feenstra, B.; Hviid, A.; Josan, V.; Mohanraj, R.; et al. Genetic regulation of gene expression in the epileptic human hippocampus. Hum. Mol. Genet. 2017, 26, 1759–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, H.; Ruppert, A.K.; Herms, S.; Wolf, C.; Mirza-Schreiber, N.; Stegle, O.; Czamara, D.; Forstner, A.J.; Sivalingam, S.; Schoch, S.; et al. Genome-wide mapping of genetic determinants influencing DNA methylation and gene expression in human hippocampus. Nat. Commun. 2017, 8, 1511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, K.J.; White, C.C.; Patel, K.; Xu, J.; Olah, M.; Replogle, J.M.; Frangieh, M.; Cimpean, M.; Winn, P.; McHenry, A.; et al. A human microglia-like cellular model for assessing the effects of neurodegenerative disease gene variants. Sci. Transl. Med. 2017, 9, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Wan, M.; Zhang, B.; Peng, Y.; Zhou, Y.; Pi, C.; Xu, X.; Ye, L.; Zhou, X.; Zheng, L. Bivalent Histone Modifications and Development. Curr. Stem Cell Res. Ther. 2018, 13, 83–90. [Google Scholar] [CrossRef]
- Ciofani, M.; Madar, A.; Galan, C.; Sellars, M.; Mace, K.; Pauli, F.; Agarwal, A.; Huang, W.; Parkhurst, C.N.; Muratet, M.; et al. A validated regulatory network for Th17 cell specification. Cell 2012, 151, 289–303. [Google Scholar] [CrossRef] [Green Version]
- Dong, C. Targeting Th17 cells in immune diseases. Cell Res. 2014, 24, 901–903. [Google Scholar] [CrossRef] [Green Version]
- Miraldi, E.R.; Pokrovskii, M.; Watters, A.; Castro, D.M.; De Veaux, N.; Hall, J.A.; Lee, J.Y.; Ciofani, M.; Madar, A.; Carriero, N.; et al. Leveraging chromatin accessibility for transcriptional regulatory network inference in T Helper 17 Cells. Genome Res. 2019, 29, 449–463. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.T.; Kanno, Y.; Cannons, J.L.; Handon, R.; Bible, P.; Elkahloun, A.G.; Anderson, S.M.; Wei, L.; Sun, H.; O’Shea, J.J.; et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity 2011, 35, 622–632. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, M.A.; Jaronen, M.; Covacu, R.; Zandee, S.E.J.; Scalisi, G.; Rothhammer, V.; Tjon, E.C.; Chao, C.C.; Kenison, J.E.; Blain, M.; et al. Environmental Control of Astrocyte Pathogenic Activities in CNS Inflammation. Cell 2019, 176, 581–596.e18. [Google Scholar] [CrossRef] [Green Version]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [Green Version]
- Eggen, B.J.; Raj, D.; Hanisch, U.K.; Boddeke, H.W. Microglial phenotype and adaptation. J. Neuroimmune Pharmacol. 2013, 8, 807–823. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, U.K.; Kettenmann, H. Microglia: Active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007, 10, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M.; Cardona, A.E. The myeloid cells of the central nervous system parenchyma. Nature 2010, 468, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Aloisi, F. Immune function of microglia. Glia 2001, 36, 165–179. [Google Scholar] [CrossRef]
- Kettenmann, H.; Ransom, B.R. Neuroglia, 2nd ed.; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Papageorgiou, I.E.; Fetani, A.F.; Lewen, A.; Heinemann, U.; Kann, O. Widespread activation of microglial cells in the hippocampus of chronic epileptic rats correlates only partially with neurodegeneration. Brain Struct. Funct. 2015, 220, 2423–2439. [Google Scholar] [CrossRef] [PubMed]
- Boche, D.; Perry, V.H.; Nicoll, J.A.R. Review: Activation patterns of microglia and their identification in the human brain. Neuropathol. Appl. Neurobiol. 2013, 39, 3–18. [Google Scholar] [CrossRef]
- Gordon, S.; Plűddemann, A. Tissue macrophage heterogeneity: Issues and prospects. Semin. Immunopathol. 2013, 35, 533–540. [Google Scholar] [CrossRef]
- Gordon, S.; Plüddemann, A.; Martinez Estrada, F. Macrophage heterogeneity in tissues: Phenotypic diversity and functions. Immunol. Rev. 2014, 262, 36–55. [Google Scholar] [CrossRef] [Green Version]
- Crotti, A.; Ransohoff, R.M. Microglial Physiology and Pathophysiology: Insights from Genome-wide Transcriptional Profiling. Immunity 2016, 44, 505–515. [Google Scholar] [CrossRef] [Green Version]
- Holtman, I.R.; Noback, M.; Bijlsma, M.; Duong, K.N.; van der Geest, M.A.; Ketelaars, P.T.; Brouwer, N.; Vainchtein, I.D.; Eggen, B.J.L.; Boddeke, H.W.G.M. Glia Open Access Database (GOAD): A comprehensive gene expression encyclopedia of glia cells in health and disease. Glia 2015, 63, 1495–1506. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.; Means, T.K.; El Khoury, J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013, 16, 1896–1905. [Google Scholar] [CrossRef] [Green Version]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gosselin, D.; Skola, D.; Coufal, N.G.; Holtman, I.R.; Schlachetzki, J.C.M.; Sajti, E.; Jaeger, B.N.; O’Connor, C.; Fitzpatrick, C.; Pasillas, M.P.; et al. An environment-dependent transcriptional network specifies human microglia identity. Science 2017, 356, 6344. [Google Scholar] [CrossRef] [Green Version]
- Hammond, T.R.; Dufort, C.; Dissing-Olesen, L.; Giera, S.; Young, A.; Wysoker, A.; Walker, A.J.; Gergits, F.; Segel, M.; Nemesh, J.; et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 2019, 50, 253–271.e6. [Google Scholar] [CrossRef] [Green Version]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef] [Green Version]
- Holtman, I.R.; Raj, D.D.; Miller, J.A.; Schaafsma, W.; Yin, Z.; Brouwer, N.; Wes, P.D.; Möller, T.; Orre, M.; Kamphuis, W.; et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co-expression meta-analysis. Acta Neuropathol. Commun. 2015, 3, 31. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Yang, Y.; Liu, L.; Liu, X. Association between five polymorphisms in vascular endothelial growth factor gene and urinary bladder cancer risk: A systematic review and meta-analysis involving 6671 subjects. Gene 2019, 698, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, X.; Boreland, A.J.; Posyton, A.; Kwan, K.; Hart, R.P.; Jiang, P. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 2020, 11, 1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashemifar, S.; Neyshabur, B.; Khan, A.A.; Xu, J. Predicting protein-protein interactions through sequence-based deep learning. Bioinforma. Oxf. Engl. 2018, 34, i802–i810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.A.; You, Z.H.; Chen, X.; Chan, K.; Luo, X. Sequence-based prediction of protein-protein interactions using weighted sparse representation model combined with global encoding. BMC Bioinform. 2016, 17, 184. [Google Scholar] [CrossRef] [Green Version]
- Miskei, M.; Horvath, A.; Vendruscolo, M.; Fuxreiter, M. Sequence-Based Prediction of Fuzzy Protein Interactions. J. Mol. Biol. 2020, 432, 2289–2303. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting protein-protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Guo, M.; Needham, C.J.; Huang, Y.; Cai, L.; Westhead, D.R. Simple sequence-based kernels do not predict protein-protein interactions. Bioinforma. Oxf. Engl. 2010, 26, 2610–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debès, C.; Wang, M.; Caetano-Anollés, G.; Gräter, F. Evolutionary optimization of protein folding. PLoS Comput. Biol. 2013, 9, e1002861. [Google Scholar] [CrossRef] [Green Version]
- Mecca, C.; Giambanco, I.; Donato, R.; Arcuri, C. Microglia and Aging: The Role of the TREM2-DAP12 and CX3CL1-CX3CR1 Axes. Int. J. Mol. Sci. 2018, 19, 318. [Google Scholar] [CrossRef] [Green Version]
- Pocock, J.M.; Kettenmann, H. Neurotransmitter receptors on microglia. Trends Neurosci. 2007, 30, 527–535. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Bolasco, G.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; et al. Synaptic pruning by microglia is necessary for normal brain development. Science 2011, 333, 1456–1458. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Paolicelli, R.C.; Sforazzini, F.; Weinhard, L.; Bolasco, G.; Pagani, F.; Vyssotski, A.L.; Bifone, A.; Gozzi, A.; Ragozzino, D.; et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat. Neurosci. 2014, 17, 400–406. [Google Scholar] [CrossRef]
- Felger, J.C.; Lotrich, F.E. Inflammatory cytokines in depression: Neurobiological mechanisms and therapeutic implications. Neuroscience 2013, 246, 199–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranjc, M.K.; Novak, M.; Pestell, R.G.; Lah, T.T. Cytokine CCL5 and receptor CCR5 axis in glioblastoma multiforme. Radiol. Oncol. 2019, 53, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Li, L.; Zhou, C.; Chen, X.; Cao, Y. Adenosine A2A receptor activation reduces brain metastasis via SDF-1/CXCR4 axis and protecting blood-brain barrier. Mol. Carcinog. 2020, 59, 390–398. [Google Scholar] [CrossRef]
- Pranzatelli, M.R. Advances in Biomarker-Guided Therapy for Pediatric- and Adult-Onset Neuroinflammatory Disorders: Targeting Chemokines/Cytokines. Front. Immunol. 2018, 9, 557. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M.; Schafer, D.; Vincent, A.; Blachère, N.E.; Bar-Or, A. Neuroinflammation: Ways in Which the Immune System Affects the Brain. Neurother. J. Am. Soc. Exp. Neurother. 2015, 12, 896–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Zou, Q.; Liao, M.; Lu, H.; Zhao, Y. A novel machine learning method for cytokine-receptor interaction prediction. Comb. Chem. High Throughput Screen. 2016, 19, 144–152. [Google Scholar] [CrossRef]
- Nath, A.; Leier, A. Improved cytokine-receptor interaction prediction by exploiting the negative sample space. BMC Bioinform. 2020, 21, 493. [Google Scholar] [CrossRef]
- Liu, T.; Shi, K.; Li, W. Deep learning methods improve linear B-cell epitope prediction. BioData Min. 2020, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.G.; Bachmann, J.; Emmerich, N.P.N.; Bachor, O.A.; Stevanović, S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Olsson, N.; Wagar, L.E.; Fast, E.; Liu, C.L.; Muftuoglu, Y.; Sworder, B.J.; Diehn, M.; Levy, R.; et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 2019, 37, 1332–1343. [Google Scholar] [CrossRef]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016, 2016, 6760830. [Google Scholar] [CrossRef] [Green Version]
- Raoufi, E.; Hemmati, M.; Eftekhari, S.; Khaksaran, K.; Mahmodi, Z.; Farajollahi, M.M.; Mohsenzadegan, M. Epitope Prediction by Novel Immunoinformatics Approach: A State-of-the-art Review. Int. J. Pept. Res. Ther. 2020, 26, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Teraguchi, S.; Saputri, D.S.; Llamas-Covarrubias, M.A.; Davila, A.; Diez, D.; Nazlica, S.A.; Rozewicki, J.; Ismanto, H.S.; Wilamowski, J.; Xie, J.; et al. Methods for sequence and structural analysis of B and T cell receptor repertoires. Comput. Struct. Biotechnol. J. 2020, 18, 2000–2011. [Google Scholar] [CrossRef] [PubMed]
- Kawakubo, H.; Matsui, Y.; Kushima, I.; Ozaki, N.; Shimamura, T. A network of networks approach for modeling interconnected brain tissue-specific networks. Bioinforma. Oxf. Engl. 2019, 35, 3092–3101. [Google Scholar] [CrossRef] [PubMed]
- Nakae, K.; Ikegaya, Y.; Ishikawa, T.; Oba, S.; Urakubo, H.; Koyama, M.; Ishii, S. A statistical method of identifying interactions in neuron-glia systems based on functional multicell Ca2+ imaging. PLoS Comput. Biol. 2014, 10, e1003949. [Google Scholar] [CrossRef] [Green Version]
- Garaschuk, O.; Verkhratsky, A. Physiology of Microglia. Methods Mol. Biol. Clifton NJ 2019, 2034, 27–40. [Google Scholar] [CrossRef]
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Tong, S.; Deng, X.; Li, J.; Qiu, P.; Wang, K. In vivo deep-brain imaging of microglia enabled by three-photon fluorescence microscopy. Opt. Lett. 2020, 45, 5271–5274. [Google Scholar] [CrossRef] [PubMed]
- Füger, P.; Hefendehl, J.K.; Veeraraghavalu, K.; Wendeln, A.C.; Schlosser, C.; Obermüller, U.; Wegenast-Braun, B.M.; Neher, J.J.; Martus, P.; Kohsaka, S.; et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 2017, 20, 1371–1376. [Google Scholar] [CrossRef]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [Green Version]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, S.; Jones, B.M.; Traniello, I.M.; Bukhari, S.A.; Halfon, M.S.; Hofmann, H.A.; Huang, S.; Katz, P.S.; Keagy, J.; Lynch, V.J.; et al. Behavior-related gene regulatory networks: A new level of organization in the brain. Proc. Natl. Acad. Sci. USA 2020, 117, 23270–23279. [Google Scholar] [CrossRef]
- Haure-Mirande, J.V.; Audrain, M.; Fanutza, T.; Kim, S.H.; Klein, W.L.; Glabe, C.; Readhead, B.; Dudley, J.T.; Blitzer, R.D.; Wang, M.; et al. Deficiency of TYROBP, an adapter protein for TREM2 and CR3 receptors, is neuroprotective in a mouse model of early Alzheimer’s pathology. Acta Neuropathol. 2017, 134, 769–788. [Google Scholar] [CrossRef] [PubMed]
- Haure-Mirande, J.V.; Wang, M.; Audrain, M.; Fanutza, T.; Kim, S.H.; Heja, S.; Readhead, B.; Dudley, J.T.; Blitzer, R.D.; Schadt, E.E.; et al. Integrative approach to sporadic Alzheimer’s disease: Deficiency of TYROBP in cerebral Aβ amyloidosis mouse normalizes clinical phenotype and complement subnetwork molecular pathology without reducing Aβ burden. Mol. Psychiatry 2019, 24, 431–446. [Google Scholar] [CrossRef]
- Barisoni, L.; Lafata, K.J.; Hewitt, S.M.; Madabhushi, A.; Balis, U.G.J. Digital pathology and computational image analysis in nephropathology. Nat. Rev. Nephrol. 2020, 16, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
- Tizhoosh, H.R.; Pantanowitz, L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J. Pathol. Inform. 2018, 9, 38. [Google Scholar] [CrossRef]
- Kan, A. Machine learning applications in cell image analysis. Immunol. Cell Biol. 2017, 95, 525–530. [Google Scholar] [CrossRef]
- Kutlu, H.; Avci, E.; Özyurt, F. White blood cells detection and classification based on regional convolutional neural networks. Med. Hypotheses 2020, 135, 109472. [Google Scholar] [CrossRef]
- Howard, C.V.; Reed, M.G. Unbiased Stereology: Three-Dimensional Measurement in Microscopy; Garland Science/BIOS Scientific Publishers: Oxon, UK, 2005. [Google Scholar]
- Valous, N.A.; Lahrmann, B.; Zhou, W.; Veltkamp, R.; Grabe, N. Multistage histopathological image segmentation of Iba1-stained murine microglias in a focal ischemia model: Methodological workflow and expert validation. J. Neurosci. Methods. 2013, 213, 250–262. [Google Scholar] [CrossRef]
- Karperien, A.L.; Jelinek, H.F. Fractal, multifractal, and lacunarity analysis of microglia in tissue engineering. Front. Bioeng. Biotechnol. 2015, 3, 51. [Google Scholar] [CrossRef] [Green Version]
- Karperien, A.; Ahammer, H.; Jelinek, H. Quantitating the subtleties of microglial morphology with fractal analysis. Front. Cell. Neurosci. 2013, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Pardon, M.C.; Agostini, A.; Faas, H.; Duan, J.; Ward, W.O.C.; Easton, F.; Auer, D.; Bai, L. Novel Methods for Microglia Segmentation, Feature Extraction, and Classification. IEEE ACM Trans. Comput. Biol. Bioinform. 2017, 14, 1366–1377. [Google Scholar] [CrossRef] [Green Version]
- Kongsui, R.; Johnson, S.J.; Graham, B.A.; Nilsson, M.; Walker, F.R. A combined cumulative threshold spectra and digital reconstruction analysis reveal structural alterations of microglia within the prefrontal cortex following low-dose LPS administration. Neuroscience 2015, 310, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ylanko, J.; Weekman, E.; Beckett, T.; Andrews, D.; McLaurin, J. Utilizing supervised machine learning to identify microglia and astrocytes in situ: Implications for large-scale image analysis and quantification. J. Neurosci. Methods. 2019, 328, 108424. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.C.G.; Carvalho-Paulo, D.; de Lima, C.M.; de Oliveira, R.B.; Santos Filho, C.; Diniz, D.G.; Bento Torres Neto, J.; Picanço-Diniz, C.W. Long-term environmental enrichment reduces microglia morphological diversity of the molecular layer of dentate gyrus. Eur. J. Neurosci. 2020, 52, 4081–4099. [Google Scholar] [CrossRef] [PubMed]
- Ohm, D.T.; Fought, A.J.; Martersteck, A.; Coventry, C.; Sridhar, J.; Gefen, T.; Weintraub, S.; Bigio, E.; Mesulam, M.M.; Rogalski, E.; et al. Accumulation of neurofibrillary tangles and activated microglia is associated with lower neuron densities in the aphasic variant of Alzheimer’s disease. Brain Pathol. Zurich Switz. 2021, 31, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Ahmady Phoulady, H.; Goldgof, D.; Hall, L.O.; Mouton, P.R. Automatic ground truth for deep learning stereology of immunostained neurons and microglia in mouse neocortex. J. Chem. Neuroanat. 2019, 98, 1–7. [Google Scholar] [CrossRef]
- Alahmari, S.S.; Goldgof, D.; Hall, L.; Phoulady, H.A.; Patel, R.H.; Mouton, P.R. Automated Cell Counts on Tissue Sections by Deep Learning and Unbiased Stereology. J. Chem. Neuroanat. 2019, 96, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Suleymanova, I.; Balassa, T.; Tripathi, S.; Molnar, C.; Saarma, M.; Sidorova, Y.; Horvath, P. A deep convolutional neural network approach for astrocyte detection. Sci. Rep. 2018, 8, 12878. [Google Scholar] [CrossRef] [PubMed]
- da Silva, B.C.G.; Tam, R.; Ferrari, R.J. Detecting cells in intravital video microscopy using a deep convolutional neural network. Comput. Biol. Med. 2021, 129, 104133. [Google Scholar] [CrossRef] [PubMed]
- Rom, S.; Zuluaga-Ramirez, V.; Dykstra, H.; Reichenbach, N.L.; Ramirez, S.H.; Persidsky, Y. Poly(ADP-ribose) polymerase-1 inhibition in brain endothelium protects the blood-brain barrier under physiologic and neuroinflammatory conditions. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2015, 35, 28–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luisi, J.; Narayanaswamy, A.; Galbreath, Z.; Roysam, B. The FARSIGHT trace editor: An open source tool for 3-D inspection and efficient pattern analysis aided editing of automated neuronal reconstructions. Neuroinformatics. 2011, 9, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Scorcioni, R.; Polavaram, S.; Ascoli, G.A. L-Measure: A web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 2008, 3, 866–876. [Google Scholar] [CrossRef]
- Lu, Y.; Carin, L.; Coifman, R.; Shain, W.; Roysam, B. Quantitative arbor analytics: Unsupervised harmonic co-clustering of populations of brain cell arbors based on L-measure. Neuroinformatics 2015, 13, 47–63. [Google Scholar] [CrossRef]
- Megjhani, M.; Rey-Villamizar, N.; Merouane, A.; Lu, Y.; Mukherjee, A.; Trett, K.; Chong, P.; Harris, C.; Shain, W.; Roysam, B. Population-scale three-dimensional reconstruction and quantitative profiling of microglia arbors. Bioinforma. Oxf. Engl. 2015, 31, 2190–2198. [Google Scholar] [CrossRef] [Green Version]
- Galbreath, Z.S. Boyer K.L.Tracing, Extracting Features, and Classifying Microglia from Volumetric Images of Brain Tissue. Master’s Thesis, Rensselaer Polytechnic Institute, New York, NY, USA, 2011. [Google Scholar]
- Wang, Y.; Narayanaswamy, A.; Tsai, C.L.; Roysam, B. A broadly applicable 3-D neuron tracing method based on open-curve snake. Neuroinformatics 2011, 9, 193–217. [Google Scholar] [CrossRef]
- Rey-Villamizar, N.; Somasundar, V.; Megjhani, M.; Xu, Y.; Lu, Y.; Padmanabhan, R.; Trett, K.; Shain, W.; Roysam, B. Large-scale automated image analysis for computational profiling of brain tissue surrounding implanted neuroprosthetic devices using Python. Front. Neuroinformatics 2014, 8, 39. [Google Scholar] [CrossRef]
- Kyriazis, A.D.; Noroozizadeh, S.; Refaee, A.; Choi, W.; Chu, L.T.; Bashir, A.; Cheng, W.H.; Zhao, R.; Namjoshi, D.R.; Salcudean, S.E.; et al. An End-to-end System for Automatic Characterization of Iba1 Immunopositive Microglia in Whole Slide Imaging. Neuroinformatics 2019, 17, 373–389. [Google Scholar] [CrossRef] [PubMed]
| Database | URL | Content |
|---|---|---|
| Sequencing | ||
| Glial heterogeneity | www.glia-network.de/index.php/databases.html (accessed on 1 July 2021) | RNA-seq databases of glial cells |
| BrainRNAseq | www.BrainRNAseq.org (accessed on 1 July 2021) | RNA-seq database of glial cells |
| ImMunoGeneTics | www.imgt.org (accessed on 1 July 2021) | sequence, genome, structure and monoclonal antibodies database |
| MSigDB, Molecular Signature DataBase | www.gsea-msigdb.org/gsea/msigdb (accessed on 1 July 2021) | Gene Set Enrichment Analysis database |
| GTEx, Genotype Tissue-Expression repository | www.gtexportal.org/home/ (accessed on 1 July 2021) | eQTL database |
| ImmPort and ImmuneSpace | www.immuneprofiling.org (accessed on 1 July 2021) | cross-essay human immunological data, including computational interface |
| IEDB, Immune Epitope DataBase | www.iedb.org/ (accessed on 1 July 2021) | antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity, and transplantation |
| ImmGen | www.immgen.org/ (accessed on 1 July 2021) | microarray dissection of gene expression and its regulation in the immune system of the mouse |
| InnateDB | www.innatedb.com/ (accessed on 1 July 2021) | genes, proteins, experimentally-verified interactions, and signaling pathways involved in the innate immune response of humans, mice, and bovines to microbial infection |
| Morphology | ||
| Neuromorph | www.neuromorpho.org (accessed on 1 July 2021) | morphology, tracing reconstructions of neurons, astroglia, and microglia |
| FARSight | farsight-toolkit.ee.uh.edu/wiki/Main_Page (accessed on 1 July 2021) | Toolkit for Python, cell arborization visualization, analysis, and quantitation applying unsupervised clustering |
| FindMyCells | www.findmycells.org/index.html (accessed on 1 July 2021) | Deep learning tool for single-cell detection |
| Assay for Transposase-Accessible Chromatin Using Sequencing (ATAC-Seq) | Method for Determining Chromatin Accessibility Across the Genome by Sequencing Regions of Open Chromatin |
| Automated Segmentation Algorithm (ASA) Stereology | Stereology with an integrated automated segmentation algorithm for cell recognition |
| Blood-brain barrier | Anatomical and functional blood vessels “seal” that keeps harmful substances from reaching the brain |
| Chromatin Immunoprecipitation and DNAseq (ChIPseq) | Chromatin Immunoprecipitation (ChIP) and next-generation sequencing (ChIPseq) explores interactions between DNA, histones, and transcription factors |
| Cytometry by Time of Flight (CyTOF) | CyTOF is a hybrid method of mass spectrometry and flow cytometry, based on isotope reporters |
| Epigenetics | Heritable changes in gene expression that take place without altering DNA sequence or modifications of the chromatin environment |
| Epigenome Wide Association studies (EWAS) | An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiable phenotype/trait |
| Expression and methylation Quantitative Trait Loci (eQTL, meQTL) | Genomic loci that explain the variation in RNA expression |
| Genome-Wide Association Studies (GWAS) | A genome-wide association study (GWAS) associates genetic variations with diseases. The method involves a population-wide genome screening and isolates genetic markers with possible disease predictive value |
| Genome-Wide Transcriptional Profiling (GWTP) | Unbiased, hypothesis-free approach that associates genetic changes with disparate states of the immune system to construct genotype-phenotype associations |
| h-iPSC organoids | Organoids are in vitro cultured three-dimensional structures that recapitulate key aspects of in vivo organs. They can be established from pluripotent stem cells (PSC) and induced adult stem cells (iPSC). The abbreviation “h-” speaks for the human PSC origin |
| Human Leukocyte Antigen (HLA) type | HLA are proteins that are located on the surface of the white blood cells and other tissues in the body. There are three general groups of HLA: HLA-A, HLA-B, and HLA-DR |
| Lacunarity and fractal dimension | The fractal dimension represents the roughness (hence texture) Lacunarity is a measure of gaps between (the fractal) objects |
| Lineage trajectories | Clonal lineage tracing of stem cells to define the outcome of differentiation |
| Microglial sensome | Proteomics, genomics, and epigenomics defining microglial reactive states |
| Microglial sensotypes | Newly-introduced categorization of microglia based on the deep sequencing profile (sensome) as a response to stimuli |
| Micro-RNAs (miRNA) | A microRNA (abbreviated miRNA) is a small single-stranded non-coding RNA molecule (containing about 22 nucleotides) found in plants, animals, and some viruses that functions in RNA silencing and post-transcriptional regulation of gene expression |
| Neuroimmunoinformatics | Immunoinformatics of the central neural system |
| Neuroinformatics | Neuroinformatics is the field that combines informatics and neuroscience. Neuroinformatics is related to neuroscience data and information processing by artificial neural networks |
| Probabilistic causal network models | More popular with the name “Bayesian networks”, probabilistic causal networks provide a mathematical model for inferring causal relationships among molecular and higher-order phenotypes. Bayesian networks represent the most popular subcategory of probabilistic causal networks |
| PU.1 | Transcription factor that activates gene expression during myeloid and B-lymphocyte development |
| SALL1 | Transcriptional regulator encoding gene allocated in microglia |
| Signature gene libraries | Cell type-specific gene clusters |
| Single-cell profile deconvolution | Predict cell-specific profiles from large cell populations using machine learning and signature gene libraries |
| T-helper cell | Helper T cell, also called the CD4+ cell, T helper cell, or helper T lymphocyte, a type of white blood cell and key mediator of the immune function |
| Ventral and dorsal h-iPSC organoids | Organoids deriving from ventral and dorsal neurotube stem cells |
| Xenografting | Xenotransplantation or heterologous transplant, the transplantation of living cells, tissues or organs from one species to another. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papageorgiou, I.; Bittner, D.; Psychogios, M.N.; Hadjidemetriou, S. Brain Immunoinformatics: A Symmetrical Link between Informatics, Wet Lab and the Clinic. Symmetry 2021, 13, 2168. https://0-doi-org.brum.beds.ac.uk/10.3390/sym13112168
Papageorgiou I, Bittner D, Psychogios MN, Hadjidemetriou S. Brain Immunoinformatics: A Symmetrical Link between Informatics, Wet Lab and the Clinic. Symmetry. 2021; 13(11):2168. https://0-doi-org.brum.beds.ac.uk/10.3390/sym13112168
Chicago/Turabian StylePapageorgiou, Ismini, Daniel Bittner, Marios Nikos Psychogios, and Stathis Hadjidemetriou. 2021. "Brain Immunoinformatics: A Symmetrical Link between Informatics, Wet Lab and the Clinic" Symmetry 13, no. 11: 2168. https://0-doi-org.brum.beds.ac.uk/10.3390/sym13112168