Does Transfusion of Red Blood Cells Impact Germline Genetic Test Results?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Red Blood Cell Unit Collection and Processing
2.2. In Vitro Experiment Calculations and Assumptions
2.3. Molecular Testing and Data Analysis
2.4. Comparison to Analytic Sensitivity of Molecular Techniques
3. Results
In Vitro Transfusion Simulations
4. Discussion
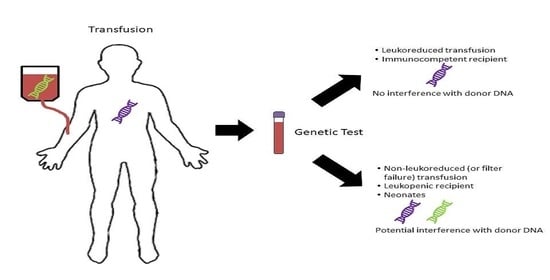
- Leukoreduced blood products are exceedingly unlikely to affect genetic testing in patients of any age. Information regarding this can be easily obtained from the institution’s blood bank or laboratory director;
- Transfusion of non-leukoreduced products can lead to appreciable amounts of donor DNA and these situations need further investigation including information such as the number of products transfused, the length of time since the last transfusion, and the clinical scenario.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gammill, H.S.; Nelson, J.L. Naturally acquired microchimerism. Int. J. Dev. Biol. 2010, 54, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Nelson, J.L. Microchimerism in human health and disease. Autoimmunity 2003, 36, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Naik, R.; Suryawanshi, H.; Gupta, N. Microchimerism: A new concept. J. Oral. Maxillofac. Pathol. 2019, 23, 311. [Google Scholar] [CrossRef] [PubMed]
- Utter, G.H.; Reed, W.F.; Lee, T.H.; Busch, M.P. Transfusion-associated microchimerism. Vox Sang. 2007, 93, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Bloch, E.M.; Jackman, R.P.; Lee, T.H.; Busch, M.P. Transfusion-associated microchimerism: The hybrid within. Transfus. Med. Rev. 2013, 27, 10–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirani, R.; Dean, M.M.; Balogh, Z.J.; Lott, N.J.; Seggie, J.; Hsu, J.M.; Taggart, S.; Maitz, P.; Survela, L.; Joseph, A.; et al. Donor white blood cell survival and cytokine profiles following red blood cell transfusion in Australian major trauma patients. Mol. Immunol. 2018, 103, 229–234. [Google Scholar] [CrossRef]
- Lee, T.H.; Paglieroni, T.; Utter, G.H.; Chafets, D.; Gosselin, R.C.; Reed, W.; Owings, J.T.; Holland, P.V.; Busch, M.P. High-level long-term white blood cell microchimerism after transfusion of leukoreduced blood components to patients resuscitated after severe traumatic injury. Transfusion 2005, 45, 1280–1290. [Google Scholar] [CrossRef]
- Utter, G.H.; Nathens, A.B.; Lee, T.H.; Reed, W.F.; Owings, J.T.; Nester, T.A.; Busch, M.P. Leukoreduction of blood transfusions does not diminish transfusion-associated microchimerism in trauma patients. Transfusion 2006, 46, 1863–1869. [Google Scholar] [CrossRef]
- Wenk, R.E.; Chiafari, P.A. DNA typing of recipient blood after massive transfusion. Transfusion 1997, 37, 1108–1110. [Google Scholar] [CrossRef]
- Jacob, R.P.; Dean, C.L.; Krummey, S.M.; Shah, Z.; Sutherland, N.; Orear, C.; Gebel, H.M.; Bray, R.A.; Sullivan, H.C. The impact of transfused blood products on deceased donor HLA typing. Hum. Immunol. 2019, 80, 976–982. [Google Scholar] [CrossRef]
- Bassuni, W.Y.; Blajchman, M.A.; Al-Moshary, M.A. Why implement universal leukoreduction? Hematol. Oncol. Stem Cell Ther. 2008, 1, 106–123. [Google Scholar] [CrossRef] [Green Version]
- Phillips, K.A.; Deverka, P.A.; Hooker, G.W.; Douglas, M.P. Genetic Test Availability And Spending: Where Are We Now? Where Are We Going? Health Aff. 2018, 37, 710–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiede, C.; Prange-Krex, G.; Freiberg-Richter, J.; Bornhäuser, M.; Ehninger, G. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant. 2000, 25, 575–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsiatis, A.C.; Norris-Kirby, A.; Rich, R.G.; Hafez, M.J.; Gocke, C.D.; Eshleman, J.R.; Murphy, K.M. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: Diagnostic and clinical implications. J. Mol. Diagn. 2010, 12, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, L.; Flaws, M. Molecular Diagnostics: Fundamentals, Methods, & Clinical Applications; F.A. Davis Company: Philadelphia, PA, USA, 2007. [Google Scholar]
- Peltonen, J.; Welsh, J.A.; Vähäkangas, K.H. Is there a role for PCR-SSCP among the methods for missense mutation detection of TP53 gene? Hum. Exp. Toxicol. 2007, 26, 9–18. [Google Scholar] [CrossRef]
- Uchiyama, Y.; Nakashima, M.; Watanabe, S.; Miyajima, M.; Taguri, M.; Miyatake, S.; Miyake, N.; Saitsu, H.; Mishima, H.; Kinoshita, A.; et al. Ultra-sensitive droplet digital PCR for detecting a low-prevalence somatic GNAQ mutation in Sturge-Weber syndrome. Sci. Rep. 2016, 6, 22985. [Google Scholar] [CrossRef] [Green Version]
- Reid, M.E.; Rios, M.; Powell, V.I.; Charles-Pierre, D.; Malavade, V. DNA from blood samples can be used to genotype patients who have recently received a transfusion. Transfusion 2000, 40, 48–53. [Google Scholar] [CrossRef]
- Gong, M.N.; Sai, Y.; Zhou, W.; Thompson, B.T.; Xu, L.L.; Christiani, D.C. Genotyping patients with recent blood transfusions. Epidemiology 2003, 14, 744–747. [Google Scholar] [CrossRef] [Green Version]
- Yomtovian, R.; Gernsheimer, T.; Assmann, S.F.; Mohandas, K.; Lee, T.H.; Kalish, L.A.; Busch, M.P. WBC reduction in RBC concentrates by prestorage filtration: Multicenter experience. Transfusion 2001, 41, 1030–1036. [Google Scholar] [CrossRef]
- Ledent, E.; Berlin, G. Inadequate white cell reduction by bedside filtration of red cell concentrates. Transfusion 1994, 34, 765–768. [Google Scholar] [CrossRef] [Green Version]
- van der Meer, P.F.; Pietersz, R.N.; Nelis, J.T.; Hinloopen, B.; Dekker, W.J.; Reesink, H.W. Six filters for the removal of white cells from red cell concentrates, evaluated at 4 degrees C and/or at room temperature. Transfusion 1999, 39, 265–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, K.J.; Mickel, M.; Braine, H.G.; Davis, K.; Enright, H.; Gernsheimer, T.; Gillespie, M.J.; Kickler, T.S.; Lee, E.J.; McCullough, J.J.; et al. White cell reduction in platelet concentrates and packed red cells by filtration: A multicenter clinical trial. The Trap Study Group. Transfusion 1995, 35, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Donegan, E.; Slichter, S.; Busch, M.P. Transient increase in circulating donor leukocytes after allogeneic transfusions in immunocompetent recipients compatible with donor cell proliferation. Blood 1995, 85, 1207–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, R.; Lee, T.H.; Wen, L.; Montalvo, L.; Schechterly, C.; Colvin, C.; Alter, H.J.; Luban, N.L.; Busch, M.P. Absence of transfusion-associated microchimerism in pediatric and adult recipients of leukoreduced and gamma-irradiated blood components. Transfusion 2012, 52, 936–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Immunocompetent Recipient | |||
|---|---|---|---|
| DONOR | LR | PLR | NLR |
| 1.2 mL (2 units) | 0.0% | 0.0% | 13.3% |
| 3.0 mL (5 units) | 0.0% | 0.1% | 24.1% |
| 10 mL (16 units) | No Amp | 1.5% | 45.1% |
| Leukopenic Recipient | |||
|---|---|---|---|
| DONOR | LR | PLR | NLR |
| 1.2 mL (2 units) | 0.0% | 6.3% | 75.9% |
| 3.0 mL (5 units) | 0.0% | 12.2% | 88.1% |
| 10 mL (16 units) | 0.0% | 27.8% | 95.7% |
| Technique | Approximate Analytic Sensitivity |
|---|---|
| Sanger Sequencing | 10–20% [14] |
| Restriction Fragment Length Polymorphism Analysis | 0.1–1% [15] |
| Pyrosequencing | 5% [14] |
| Next Generation Sequencing (germline) | 5–10% * |
| Next Generation Sequencing (somatic) | 5% ** |
| Real-Time PCR with Melt Curve Analysis | 1–10% [14] |
| Single-Stranded Conformational Polymorphism Analysis with Denaturing High Performance Liquid Chromatography | 1–5% [16] |
| Microsatellite Short Tandem Repeat Analysis by Capillary Electrophoresis | 1% *** |
| Amplification Refractory Mutation System Analysis | 0.05–0.1% [16] |
| Droplet Digital PCR | 0.25% [17] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
DiGuardo, M.A.; Kester, S.J.; Mahaffey, V.J.; Hammel, S.A.; Heaser, K.K.; Hofich, C.D.; Tauscher, C.D.; Kerr, S.E.; Oliveira, J.L.; Jacob, E.K.; et al. Does Transfusion of Red Blood Cells Impact Germline Genetic Test Results? J. Pers. Med. 2020, 10, 268. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm10040268
DiGuardo MA, Kester SJ, Mahaffey VJ, Hammel SA, Heaser KK, Hofich CD, Tauscher CD, Kerr SE, Oliveira JL, Jacob EK, et al. Does Transfusion of Red Blood Cells Impact Germline Genetic Test Results? Journal of Personalized Medicine. 2020; 10(4):268. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm10040268
Chicago/Turabian StyleDiGuardo, Maggie A., Sarah J. Kester, Victor J. Mahaffey, Scott A. Hammel, Katelyn K. Heaser, Christopher D. Hofich, Craig D. Tauscher, Sarah E. Kerr, Jennifer L. Oliveira, Eapen K. Jacob, and et al. 2020. "Does Transfusion of Red Blood Cells Impact Germline Genetic Test Results?" Journal of Personalized Medicine 10, no. 4: 268. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm10040268