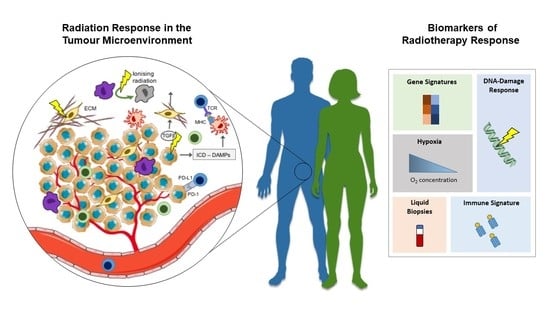
Radiation Response in the Tumour Microenvironment: Predictive Biomarkers and Future Perspectives
Abstract
:1. Introduction
2. Radiation Response in the Tumour Microenvironment
2.1. Tumour Immune Microenvironment
2.2. Cancer-Associated Fibroblasts
2.3. Tumour Vasculature
3. Predictive Biomarkers of Radiation Response
3.1. Gene Signatures of Radiation Sensitivity
3.2. DNA Damage Response Biomarkers
3.3. Hypoxia Biomarkers
3.4. Liquid Biopsies
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernier, J.; Hall, E.J.; Giaccia, A. Radiation oncology: A century of achievements. Nat. Rev. Cancer 2004, 4, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Barcellos-Hoff, M.H.; Park, C.; Wright, E.G. Radiation and the microenvironment—tumorigenesis and therapy. Nat. Rev. Cancer 2005, 5, 867–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Barker, H.E.; Paget, J.T.; Khan, A.A.; Harrington, K.J. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef]
- Bratman, S.V.; Milosevic, M.F.; Liu, F.F.; Haibe-Kains, B. Genomic biomarkers for precision radiation medicine. Lancet Oncol. 2017, 18, e238. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.G.; Berglund, A.; Schell, M.J.; Mihaylov, I.; Fulp, W.J.; Yue, B.; Welsh, E.; Caudell, J.J.; Ahmed, K.; Strom, T.S.; et al. A genome-based model for adjusting radiotherapy dose (GARD): A retrospective, cohort-based study. Lancet Oncol. 2017, 18, 202–211. [Google Scholar] [CrossRef]
- Ni, J.; Bucci, J.; Malouf, D.; Knox, M.; Graham, P.; Li, Y. Exosomes in Cancer Radioresistance. Front. Oncol. 2019, 9, 869. [Google Scholar] [CrossRef] [Green Version]
- Szatmári, T.; Hargitai, R.; Sáfrány, G.; Lumniczky, K. Extracellular Vesicles in Modifying the Effects of Ionizing Radiation. Int. J. Mol. Sci. 2019, 20, 5527. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F. The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin. Oncol. 2015, 42, 663–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front. Immunol. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Q.; Zhang, H.; Zheng, J.; Zhang, L. Turning Cold into Hot: Firing up the Tumor Microenvironment. Trends Cancer 2020, 6, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Dudzinski, S.O.; Cameron, B.D.; Wang, J.; Rathmell, J.C.; Giorgio, T.D.; Kirschner, A.N. Combination immunotherapy and radiotherapy causes an abscopal treatment response in a mouse model of castration resistant prostate cancer. J. Immunother. Cancer 2019, 7, 218. [Google Scholar] [CrossRef] [Green Version]
- Reynders, K.; Illidge, T.; Siva, S.; Chang, J.Y.; De Ruysscher, D. The abscopal effect of local radiotherapy: Using immunotherapy to make a rare event clinically relevant. Cancer Treat. Rev. 2015, 41, 503–510. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Le, T.Q.; Massarelli, E.; Hendifar, A.E.; Tuli, R. Radiation therapy and PD-1/PD-L1 blockade: The clinical development of an evolving anticancer combination. J. Immunother. Cancer 2018, 6, 46. [Google Scholar] [CrossRef]
- Roger, A.; Finet, A.; Boru, B.; Beauchet, A.; Mazeron, J.J.; Otzmeguine, Y.; Blom, A.; Longvert, C.; de Maleissye, M.F.; Fort, M.; et al. Efficacy of combined hypo-fractionated radiotherapy and anti-PD-1 monotherapy in difficult-to-treat advanced melanoma patients. Oncoimmunology 2018, 7, e1442166. [Google Scholar] [CrossRef]
- Demaria, S.; Kawashima, N.; Yang, A.M.; Devitt, M.L.; Babb, J.S.; Allison, J.P.; Formenti, S.C. Immune-mediated inhibition of metastases after treatment with local radiation and CTLA-4 blockade in a mouse model of breast cancer. Clin. Cancer Res. 2005, 11, 728–734. [Google Scholar] [PubMed]
- Postow, M.A.; Callahan, M.K.; Barker, C.A.; Yamada, Y.; Yuan, J.; Kitano, S.; Mu, Z.; Rasalan, T.; Adamow, M.; Ritter, E.; et al. Immunologic correlates of the abscopal effect in a patient with melanoma. N. Engl. J. Med. 2012, 366, 925–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, R.A.; Wilhite, T.J.; Balboni, T.A.; Alexander, B.M.; Spektor, A.; Ott, P.A.; Ng, A.K.; Hodi, F.S.; Schoenfeld, J.D. A systematic evaluation of abscopal responses following radiotherapy in patients with metastatic melanoma treated with ipilimumab. Oncoimmunology 2015, 4, e1046028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garelli, E.; Rittmeyer, A.; Putora, P.M.; Glatzer, M.; Dressel, R.; Andreas, S. Abscopal effect in lung cancer: Three case reports and a concise review. Immunotherapy 2019, 11, 1445–1461. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Xu, Y.; Chen, X.; Liu, J.; Weng, Y.; Zhuang, Q.; Lin, F.; Huang, Z.; Wu, S.; Ding, J.; et al. Radiation-induced small extracellular vesicles as "carriages" promote tumor antigen release and trigger antitumor immunity. Theranostics 2020, 10, 4871–4884. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, M.E.; Vanpouille-Box, C.; Melero, I.; Formenti, S.C.; Demaria, S. Immunological Mechanisms Responsible for Radiation-Induced Abscopal Effect. Trends Immunol. 2018, 39, 644–655. [Google Scholar] [CrossRef]
- Huang, R.X.; Zhou, P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Golden, E.B.; Apetoh, L. Radiotherapy and immunogenic cell death. Semin. Radiat. Oncol. 2015, 25, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Golden, E.B.; Frances, D.; Pellicciotta, I.; Demaria, S.; Helen Barcellos-Hoff, M.; Formenti, S.C. Radiation fosters dose-dependent and chemotherapy-induced immunogenic cell death. Oncoimmunology 2014, 3, e28518. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Bode, B.; Wenger, R.H.; Lehmann, K.; Sartori, A.A.; Moch, H.; Knuth, A.; Boehmer, L.; Broek, M. γ-Radiation promotes immunological recognition of cancer cells through increased expression of cancer-testis antigens in vitro and in vivo. PLoS ONE 2011, 6, e28217. [Google Scholar] [CrossRef]
- Deng, L.; Liang, H.; Xu, M.; Yang, X.; Burnette, B.; Arina, A.; Li, X.D.; Mauceri, H.; Beckett, M.; Darga, T.; et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014, 41, 843–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Qu, J.; Sun, Y.; Wang, J.; Liu, X.; Wang, F.; Zhang, H.; Wang, W.; Ma, X.; Gao, X.; et al. Prognostic significance of tumor-associated macrophages in breast cancer: A meta-analysis of the literature. Oncotarget 2017, 8, 30576–30586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuri, P.; Shigemura, K.; Kitagawa, K.; Hadibrata, E.; Risan, M.; Zulfiqqar, A.; Soeroharjo, I.; Hendri, A.Z.; Danarto, R.; Ishii, A.; et al. Increased tumor-associated macrophages in the prostate cancer microenvironment predicted patients’ survival and responses to androgen deprivation therapies in Indonesian patients cohort. Prostate Int. 2020, 8, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Klug, F.; Prakash, H.; Huber, P.E.; Seibel, T.; Bender, N.; Halama, N.; Pfirschke, C.; Voss, R.H.; Timke, C.; Umansky, L.; et al. Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell 2013, 24, 589–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiao, S.L.; Ruffell, B.; DeNardo, D.G.; Faddegon, B.A.; Park, C.C.; Coussens, L.M. TH2-Polarized CD4(+) T Cells and Macrophages Limit Efficacy of Radiotherapy. Cancer Immunol. Res. 2015, 3, 518–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Escamilla, J.; Mok, S.; David, J.; Priceman, S.; West, B.; Bollag, G.; McBride, W.; Wu, L. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer Res. 2013, 73, 2782–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stary, V.; Wolf, B.; Unterleuthner, D.; List, J.; Talic, M.; Laengle, J.; Beer, A.; Strobl, J.; Stary, G.; Dolznig, H.; et al. Short-course radiotherapy promotes pro-inflammatory macrophages via extracellular vesicles in human rectal cancer. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]
- Straub, J.M.; New, J.; Hamilton, C.D.; Lominska, C.; Shnayder, Y.; Thomas, S.M. Radiation-induced fibrosis: Mechanisms and implications for therapy. J. Cancer Res. Clin. Oncol. 2015, 141, 1985–1994. [Google Scholar] [CrossRef] [Green Version]
- Dancea, H.C.; Shareef, M.M.; Ahmed, M.M. Role of Radiation-induced TGF-beta Signaling in Cancer Therapy. Mol. Cell. Pharmacol. 2009, 1, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganguly, D.; Chandra, R.; Karalis, J.; Teke, M.; Aguilera, T.; Maddipati, R.; Wachsmann, M.B.; Ghersi, D.; Siravegna, G.; Zeh, H.J., 3rd; et al. Cancer-Associated Fibroblasts: Versatile Players in the Tumor Microenvironment. Cancers (Basel) 2020, 12, 2652. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, Y.; Tan, Y.; Wei, Q.; Yu, W. Cancer-associated fibroblasts in radiotherapy: Challenges and new opportunities. Cell Commun. Signal. 2019, 17, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinde, M.T.; Vik, J.; Camilio, K.A.; Martinez-Zubiaurre, I.; Hellevik, T. Ionizing radiation abrogates the pro-tumorigenic capacity of cancer-associated fibroblasts co-implanted in xenografts. Sci. Rep. 2017, 7, 46714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamochi, N.; Nakashima, M.; Aoki, S.; Uchihashi, K.; Sugihara, H.; Toda, S.; Kudo, S. Irradiated fibroblast-induced bystander effects on invasive growth of squamous cell carcinoma under cancer-stromal cell interaction. Cancer Sci. 2008, 99, 2417–2427. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Z.; Zhou, L.; Hu, L.; Yin, C.; Qing, D.; Huang, S.; Cai, X.; Chen, Y. Cancer associated fibroblasts-derived exosomes contribute to radioresistance through promoting colorectal cancer stem cells phenotype. Exp. Cell Res. 2020, 391, 111956. [Google Scholar] [CrossRef]
- Colton, M.; Cheadle, E.J.; Honeychurch, J.; Illidge, T.M. Reprogramming the tumour microenvironment by radiotherapy: Implications for radiotherapy and immunotherapy combinations. Radiat. Oncol. 2020, 15, 254. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M. Radiation Damage to Tumor Vasculature Initiates a Program That Promotes Tumor Recurrences. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 734–744. [Google Scholar] [CrossRef]
- Castle, K.D.; Kirsch, D.G. Establishing the Impact of Vascular Damage on Tumor Response to High-Dose Radiation Therapy. Cancer Res. 2019, 79, 5685–5692. [Google Scholar] [CrossRef] [Green Version]
- Arnold, K.M.; Flynn, N.J.; Raben, A.; Romak, L.; Yu, Y.; Dicker, A.P.; Mourtada, F.; Sims-Mourtada, J. The Impact of Radiation on the Tumor Microenvironment: Effect of Dose and Fractionation Schedules. Cancer Growth Metastasis 2018, 11, 1179064418761639. [Google Scholar] [CrossRef]
- Park, H.J.; Griffin, R.J.; Hui, S.; Levitt, S.H.; Song, C.W. Radiation-induced vascular damage in tumors: Implications of vascular damage in ablative hypofractionated radiotherapy (SBRT and SRS). Radiat. Res. 2012, 177, 311–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riva, M.; Wouters, R.; Nittner, D.; Ceuster, J.; Sterpin, E.; Giovannoni, R.; Himmelreich, U.; Gsell, W.; Van Ranst, M.; Coosemans, A. Radiation dose-escalation and dose-fractionation modulate the immune microenvironment, cancer stem cells and vasculature in experimental high-grade gliomas. J. Neurosurg. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Potiron, V.; Clément-Colmou, K.; Jouglar, E.; Pietri, M.; Chiavassa, S.; Delpon, G.; Paris, F.; Supiot, S. Tumor vasculature remodeling by radiation therapy increases doxorubicin distribution and efficacy. Cancer Lett. 2019, 457, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kouam, P.N.; Rezniczek, G.A.; Adamietz, I.A.; Bühler, H. Ionizing radiation increases the endothelial permeability and the transendothelial migration of tumor cells through ADAM10-activation and subsequent degradation of VE-cadherin. BMC Cancer 2019, 19, 958. [Google Scholar] [CrossRef] [PubMed]
- Maeda, A.; Chen, Y.; Bu, J.; Mujcic, H.; Wouters, B.G.; DaCosta, R.S. In Vivo Imaging Reveals Significant Tumor Vascular Dysfunction and Increased Tumor Hypoxia-Inducible Factor-1α Expression Induced by High Single-Dose Irradiation in a Pancreatic Tumor Model. Int. J. Radiat. Oncol. Biol. Phys. 2017, 97, 184–194. [Google Scholar] [CrossRef]
- Banerjee, D.; Barton, S.M.; Grabham, P.W.; Rumeld, A.L.; Okochi, S.; Street, C.; Kadenhe-Chiweshe, A.; Boboila, S.; Yamashiro, D.J.; Connolly, E.P. High-Dose Radiation Increases Notch1 in Tumor Vasculature. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 857–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapleton, S.; Jaffray, D.; Milosevic, M. Radiation effects on the tumor microenvironment: Implications for nanomedicine delivery. Adv. Drug Deliv. Rev. 2017, 109, 119–130. [Google Scholar] [CrossRef]
- Jensen, M.B.; Lænkholm, A.V.; Balslev, E.; Buckingham, W.; Ferree, S.; Glavicic, V.; Dupont Jensen, J.; Søegaard Knoop, A.; Mouridsen, H.T.; Nielsen, D.; et al. The Prosigna 50-gene profile and responsiveness to adjuvant anthracycline-based chemotherapy in high-risk breast cancer patients. NPJ Breast Cancer 2020, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Kim, S.C.; Kim, S.J.; Park, C.H.; Jeung, H.C.; Kim, Y.B.; Ahn, J.B.; Chung, H.C.; Rha, S.Y. Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells. BMC Genom. 2012, 13, 348. [Google Scholar] [CrossRef] [Green Version]
- Tramm, T.; Mohammed, H.; Myhre, S.; Kyndi, M.; Alsner, J.; Børresen-Dale, A.L.; Sørlie, T.; Frigessi, A.; Overgaard, J. Development and validation of a gene profile predicting benefit of postmastectomy radiotherapy in patients with high-risk breast cancer: A study of gene expression in the DBCG82bc cohort. Clin. Cancer Res. 2014, 20, 5272–5280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speers, C.; Zhao, S.; Liu, M.; Bartelink, H.; Pierce, L.J.; Feng, F.Y. Development and Validation of a Novel Radiosensitivity Signature in Human Breast Cancer. Clin. Cancer Res. 2015, 21, 3667–3677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.G.; Chang, S.L.; Spratt, D.E.; Erho, N.; Yu, M.; Ashab, H.A.; Alshalalfa, M.; Speers, C.; Tomlins, S.A.; Davicioni, E.; et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: A matched, retrospective analysis. Lancet Oncol. 2016, 17, 1612–1620. [Google Scholar] [CrossRef]
- Liu, J.; Han, M.; Yue, Z.; Dong, C.; Wen, P.; Zhao, G.; Wu, L.; Xia, J.; Bin, Y. Prediction of Radiosensitivity in Head and Neck Squamous Cell Carcinoma Based on Multiple Omics Data. Front. Genet. 2020, 11, 960. [Google Scholar] [CrossRef] [PubMed]
- Piening, B.D.; Wang, P.; Subramanian, A.; Paulovich, A.G. A radiation-derived gene expression signature predicts clinical outcome for breast cancer patients. Radiat. Res. 2009, 171, 141–154. [Google Scholar] [CrossRef]
- Oh, D.S.; Cheang, M.C.; Fan, C.; Perou, C.M. Radiation-induced gene signature predicts pathologic complete response to neoadjuvant chemotherapy in breast cancer patients. Radiat. Res. 2014, 181, 193–207. [Google Scholar] [CrossRef]
- Eustace, A.; Mani, N.; Span, P.N.; Irlam, J.J.; Taylor, J.; Betts, G.N.; Denley, H.; Miller, C.J.; Homer, J.J.; Rojas, A.M.; et al. A 26-gene hypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin. Cancer Res. 2013, 19, 4879–4888. [Google Scholar] [CrossRef] [Green Version]
- Toustrup, K.; Sørensen, B.S.; Lassen, P.; Wiuf, C.; Alsner, J.; Overgaard, J. Gene expression classifier predicts for hypoxic modification of radiotherapy with nimorazole in squamous cell carcinomas of the head and neck. Radiother. Oncol. 2012, 102, 122–129. [Google Scholar] [CrossRef]
- Li, B.; Shi, X.B.; Nori, D.; Chao, C.K.; Chen, A.M.; Valicenti, R.; White Rde, V. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate 2011, 71, 567–574. [Google Scholar] [CrossRef]
- Li, A.L.; Chung, T.S.; Chan, Y.N.; Chen, C.L.; Lin, S.C.; Chiang, Y.R.; Lin, C.H.; Chen, C.C.; Ma, N. microRNA expression pattern as an ancillary prognostic signature for radiotherapy. J. Transl. Med. 2018, 16, 341. [Google Scholar] [CrossRef]
- Cui, Y.; Li, B.; Pollom, E.L.; Horst, K.C.; Li, R. Integrating Radiosensitivity and Immune Gene Signatures for Predicting Benefit of Radiotherapy in Breast Cancer. Clin. Cancer Res. 2018, 24, 4754–4762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; He, G.; Hou, M.; Chen, L.; Chen, S.; Xu, A.; Fu, Y. Cell Cycle Regulation by Alternative Polyadenylation of CCND1. Sci. Rep. 2018, 8, 6824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goel, H.L.; Mercurio, A.M. VEGF targets the tumour cell. Nat. Rev. Cancer 2013, 13, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Gorlia, T.; Erridge, S.C.; Perry, J.; Hong, Y.K.; Aldape, K.D.; Lhermitte, B.; Pietsch, T.; Grujicic, D.; et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 1100–1108. [Google Scholar] [CrossRef] [Green Version]
- Gvozdenovic, A.; Boro, A.; Meier, D.; Bode-Lesniewska, B.; Born, W.; Muff, R.; Fuchs, B. Targeting αvβ3 and αvβ5 integrins inhibits pulmonary metastasis in an intratibial xenograft osteosarcoma mouse model. Oncotarget 2016, 7, 55141–55154. [Google Scholar] [CrossRef] [PubMed]
- Noal, S.; Levy, C.; Hardouin, A.; Rieux, C.; Heutte, N.; Ségura, C.; Collet, F.; Allouache, D.; Switsers, O.; Delcambre, C.; et al. One-year longitudinal study of fatigue, cognitive functions, and quality of life after adjuvant radiotherapy for breast cancer. Int J. Radiat. Oncol. Biol. Phys. 2011, 81, 795–803. [Google Scholar] [CrossRef]
- Chen, X.; Zheng, J.; Zhuo, M.L.; Zhang, A.; You, Z. A six-gene-based signature for breast cancer radiotherapy sensitivity estimation. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Hu, Y.; Guo, M. Synthetic lethality strategies: Beyond BRCA1/2 mutations in pancreatic cancer. Cancer Sci. 2020, 111, 3111–3121. [Google Scholar] [CrossRef]
- Reichert, Z.R.; Wahl, D.R.; Morgan, M.A. Translation of Targeted Radiation Sensitizers into Clinical Trials. Semin. Radiat. Oncol. 2016, 26, 261–270. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, P.; Anderson, S.J.; Bear, H.D.; Geyer, C.E.; Kahlenberg, M.S.; Robidoux, A.; Margolese, R.G.; Hoehn, J.L.; Vogel, V.G.; Dakhil, S.R.; et al. Preoperative chemotherapy: Updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J. Clin. Oncol. 2008, 26, 778–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; West, C.M. Hypoxia gene expression signatures as predictive biomarkers for personalising radiotherapy. Br. J. Radiol. 2019, 92, 20180036. [Google Scholar] [CrossRef] [PubMed]
- Buffa, F.M.; Harris, A.L.; West, C.M.; Miller, C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer 2010, 102, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Toustrup, K.; Sørensen, B.S.; Nordsmark, M.; Busk, M.; Wiuf, C.; Alsner, J.; Overgaard, J. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res. 2011, 71, 5923–5931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordsmark, M.; Overgaard, M.; Overgaard, J. Pretreatment oxygenation predicts radiation response in advanced squamous cell carcinoma of the head and neck. Radiother. Oncol. 1996, 41, 31–39. [Google Scholar] [CrossRef]
- Nordsmark, M.; Overgaard, J. A confirmatory prognostic study on oxygenation status and loco-regional control in advanced head and neck squamous cell carcinoma treated by radiation therapy. Radiother. Oncol. 2000, 57, 39–43. [Google Scholar] [CrossRef]
- Małachowska, B.; Tomasik, B.; Stawiski, K.; Kulkarni, S.; Guha, C.; Chowdhury, D.; Fendler, W. Circulating microRNAs as Biomarkers of Radiation Exposure: A Systematic Review and Meta-Analysis. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 390–402. [Google Scholar] [CrossRef] [Green Version]
- Asakura, K.; Kadota, T.; Matsuzaki, J.; Yoshida, Y.; Yamamoto, Y.; Nakagawa, K.; Takizawa, S.; Aoki, Y.; Nakamura, E.; Miura, J.; et al. A miRNA-based diagnostic model predicts resectable lung cancer in humans with high accuracy. Commun. Biol. 2020, 3, 134. [Google Scholar] [CrossRef] [Green Version]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.H.; Hou, R.; Pan, Y.; Gao, Y.; Yang, Y.; Tian, W.; Zhu, Y.B. A five-microRNA signature for individualized prognosis evaluation and radiotherapy guidance in patients with diffuse lower-grade glioma. J. Cell Mol. Med. 2020, 24, 7504–7514. [Google Scholar] [CrossRef]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zedan, A.H.; Hansen, T.F.; Assenholt, J.; Madsen, J.S.; Osther, P.J.S. Circulating miRNAs in localized/locally advanced prostate cancer patients after radical prostatectomy and radiotherapy. Prostate 2019, 79, 425–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, K.C.; Cooperberg, M.R. Biomarkers in prostate cancer surveillance and screening: Past, present, and future. Ther. Adv. Urol. 2013, 5, 318–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kirkby, K.J.; Kirkby, N.F.; Burnet, N.G.; Owen, H.; Mackay, R.I.; Crellin, A.; Green, S. Heavy charged particle beam therapy and related new radiotherapy technologies: The clinical potential, physics and technical developments required to deliver benefit for patients with cancer. Br. J. Radiol. 2020, 93, 20200247. [Google Scholar] [CrossRef]
- Lupu-Plesu, M.; Claren, A.; Martial, S.; N’Diaye, P.D.; Lebrigand, K.; Pons, N.; Ambrosetti, D.; Peyrottes, I.; Feuillade, J.; Hérault, J.; et al. Effects of proton versus photon irradiation on (lymph)angiogenic, inflammatory, proliferative and anti-tumor immune responses in head and neck squamous cell carcinoma. Oncogenesis 2017, 6, e354. [Google Scholar] [CrossRef]
- Durante, M.; Formenti, S. Harnessing radiation to improve immunotherapy: Better with particles? Br. J. Radiol. 2020, 93, 20190224. [Google Scholar] [CrossRef]

| Year | Cancer Type | Biomarker | Results | Ref | |
|---|---|---|---|---|---|
| Gene signatures | 2012 | NCI-60 human tumor cell lines screen | A 31-gene signature developed from meta-analysis of microarray data correlated with clonogenic assay data to identify radiosensitive or radioresistant cells | Genes involved in cell cycle progression (CCNA2, CDK6, CCND1) and DNA damage repair were associated with increased radiosensitivity | [60] |
| 2014 | Breast cancer | A 7-gene signature applied to the Danish Breast Cancer Cooperative Group (DBCG82bc) cohort to stratify patients into either high-risk locoregional recurrence (LRR) or low-risk LRR | Identified that post-mastectomy RT would benefit only those identified as high risk, providing no benefit to low-risk patients | [61] | |
| 2015 | Breast cancer | Radiation sensitivity gene signature developed from correlating radiation sensitivity (SF2) of a panel of breast cancer models against gene expression changes | Gene signature significantly predicted loco-regional recurrence; beating all clinicopathologic features used in clinical practice | [62] | |
| 2016 | Prostate cancer | A 24-gene signature applied to prostate cancer patients who had undergone radical prostatectomy to identify those most likely to benefit from postoperative radiotherapy | Retrospective analysis identified that those patients with a high PROTOS (post-operative radiation therapy outcomes score), indicative of radiation sensitive tumours, were less likely to develop metastasis at 10 years post-RT. In the low PROTOS score group, radiotherapy proved detrimental | [63] | |
| 2020 | HNSCC | A 12-gene signature | Classified patients with a higher radiosensitivity for whom RT would be beneficial and could predict overall survival. | [64] | |
| DNA-damage response | 2010 | Breast cancer | Gene expression signature associated with DDR, correlated against publicly available breast cancer microarray data | DDR-associated genes induced by radiation correlated positively with those who responded favourably to radiation treatment | [65] |
| 2014 | Breast cancer | Radiation-induced 30-gene DDR signature | Gene signature was capable of discriminating between breast cancer patients likely to achieve a pathological complete response (pCR) to neoadjuvant chemotherapy and poor-responding patients | [66] | |
| Hypoxia | 2013 | Laryngeal cancer | A 26-hypoxia gene signature | Could predict those patients receiving RT for whom hypoxia-modifying ARCON (accelerated radiotherapy with carbogen and nicotinamide) therapy would be of benefit | [67] |
| 2012 | HNSCC | A 15-gene hypoxia signature | Classified patients who would benefit from combining RT with hypoxia modification (nimorazole) | [68] | |
| Liquid biopsies | 2011 | Prostate cancer | Altered miRNA expression: developed through screening of miRNAs in prostate cancer cells (LNCaP) in response to RT | Suppressed miR-221 expression linked with increased radiation sensitivity: data subsequently correlated in clinical datasets where low serum levels of miR-221 are indicative of low-risk prostate cancer | [69] |
| 2018 | Nonmetastatic rectal cancer and head and neck cancers | miRNA expression rations: prediction classifier | The expressions of three miRNAs—miR-374a-5p, miR-342-5p and miR-519d-3p—were significantly different between responsive and poor-responsive RT groups. miRNA classifier successfully predicted radiotherapy outcomes | [70] | |
| Immune signature | 2018 | Breast cancer | Combined radiation sensitivity (RS) gene signature with an antigen-presentation (AP) immune signature | Both RS and AP signatures capable of predicting increased disease specific survival (DSS) in patients identified with either radio-sensitive or immune-effective tumours | [71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byrne, N.M.; Tambe, P.; Coulter, J.A. Radiation Response in the Tumour Microenvironment: Predictive Biomarkers and Future Perspectives. J. Pers. Med. 2021, 11, 53. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11010053
Byrne NM, Tambe P, Coulter JA. Radiation Response in the Tumour Microenvironment: Predictive Biomarkers and Future Perspectives. Journal of Personalized Medicine. 2021; 11(1):53. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11010053
Chicago/Turabian StyleByrne, Niall M., Prajakta Tambe, and Jonathan A. Coulter. 2021. "Radiation Response in the Tumour Microenvironment: Predictive Biomarkers and Future Perspectives" Journal of Personalized Medicine 11, no. 1: 53. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11010053