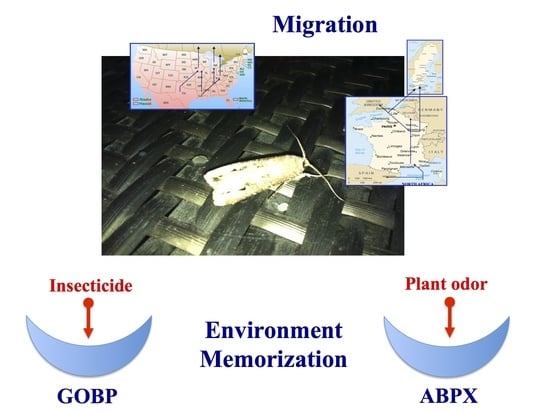
Interpopulational Variations of Odorant-Binding Protein Expression in the Black Cutworm Moth, Agrotis ipsilon
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Insect Populations
2.2. Biochemical Analysis
2.3. One-Step Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
- AipsABPX-1s/5′-ATGGCGGAGCTGGCGCGC-3′,
- AipsABPX-1as/5′-TATGAGGAAGTACTCGGCCTT-3′ (#AY301981);
- AipsPBP1s/5′-TCGCAGGAAATCATGAA-3′,
- AipsPBP1as/5′-AACTTCGGCCAAGACTTCG-3′ (#AY301985);
- AipsPBP2s/5′-TCGCAGGAGGTGGTCGCC-3′,
- AipsPBP2as/5′-CTATACGGCCGTCATGAT-3′ (#AY301986);
- AipsGOBP2s/5′-GCATATTATAGCGCACCCC-3′,
3. Results
3.1. Interpopulational Variations of A. ipsilon Antennal Protein Profiles
3.2. Interpopulational Variations of A. ipsilon OBP-RNA Levels
3.3. Interpopulational Variations of A. ipsilon p38 (Calreticulin)
4. Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Saxena, K.N.; Schoohoven, L.M. Induction of orientational and feeding preferences in Manduca sexta larvae for different food sources. Entomol. Exp. Appl. 1982, 32, 173–180. [Google Scholar] [CrossRef]
- Boer, G.; Hanson, F.E. Food plant selection and induction of feeding preference among host and non-host plants in larvae of the tobacco hornworm Manduca sexta. Entomol. Exp. Appl. 1984, 35, 177–195. [Google Scholar] [CrossRef]
- Anderson, P.; Hilker, M.; Löfqvist, J. Larval diet influence on oviposition behaviour in Spodoptera littoralis. Entomol. Exp. Appl. 1995, 74, 71–82. [Google Scholar] [CrossRef]
- Cunningham, J.P.; Moore, C.J.; Zalucki, M.P.; West, S.A. Learning, odour preference and flower foraging in moths. J. Exp. Biol. 2004, 207, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petit, C.; Le Ru, B.; Dupas, S.; Frérot, B.; Ahuya, P.; Kaiser-Arnauld, L.; Harry, M.; Calatayud, P.A. Influence of dietary experience on the induction of preference of adult moths and larvae for a new olfactory cue. PLoS ONE 2015, 10, e0136169. [Google Scholar] [CrossRef]
- Pickett, J.A.; Wadhams, L.J.; Woodcock, C.M. Insect supersense: Mate and host location by insects as model system for exploiting olfactory interactions. Biochemist 1998, 20, 8–13. [Google Scholar]
- Deisig, N.; Dupuy, F.; Anton, S.; Renou, M. Responses to pheromones in a complex odor world: Sensory processing and behavior. Insects 2014, 5, 399–422. [Google Scholar] [CrossRef]
- Heinbockel, T.; Kaissling, K.E. Variability of olfactory receptor neurons responses of female silkmoths (Bombyx mori L.) to benzoic acid and (+) linalool. J. Insect Physiol. 1996, 42, 565–578. [Google Scholar] [CrossRef]
- Kaissling, K.E. Responses of insect olfactory neurons to single pheromone molecules. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; Volume 2, Chapter 1; pp. 1–27. [Google Scholar]
- Røstelien, T. Recognition of plant odor information in moths. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; Volume 2, Chapter 3; pp. 49–91. [Google Scholar]
- Steinbrecht, R.A. Functional morphology of pheromone-sensitive sensilla. In Pheromone Biochemistry; Prestwich, G.D., Blomquist, G.J., Eds.; Academic Press: Orlando, FL, USA, 1987; pp. 353–384. [Google Scholar]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Vogt, R.G. Biochemical diversity of odor detection:OBPs, ODEs and SNMPs. In Insect Pheromone Biochemistry and Molecular Biology; Blomquist, G.J., Vogt, R.G., Eds.; Academic Press: New York, NY, USA, 2003; Chapter 14; pp. 391–445. [Google Scholar]
- Vogt, R.G. Molecular basis of pheromone detection in insects. In Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology; Gilbert, L.I., Iatrou, K., Gill, S., Eds.; Elsevier: London, UK, 2005; Volume 282, pp. 753–804. [Google Scholar]
- Picimbon, J.F. Synthesis of odorant reception-suppressing agents, Odorant-Binding Proteins (OBPs) and Chemosensory Proteins (CSPs): Molecular targets for pest management. In Biopesticides of Plant Origin; Regnault-Roger, C., Philogène, B., Vincent, C., Eds.; Intercept-Lavoisier: Paris, France; Hampshire, UK; Secaucus, NJ, USA, 2005; pp. 245–266. [Google Scholar]
- Picimbon, J.F. Olfactory Concepts of Insect Control-Alternative to Insecticides; Springer Nature Switzerland AG: Cham, Switzerland, 2019; Volume 2, p. 373. [Google Scholar]
- Steinbrecht, R.A.; Laue, M.; Ziegelberger, G. Immunolocalization of pheromone-binding-protein and general odorant-binding protein in olfactory sensilla of the silk moths Antheraea and Bombyx. Cell Tissue Res. 1995, 282, 203–217. [Google Scholar] [CrossRef]
- Vogt, R.G.; Rogers, M.E.; Franco, M.D.; Sun, M. A comparative study of odorant binding protein genes: Differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera). J. Exp. Biol. 2002, 205, 719–744. [Google Scholar] [PubMed]
- Vogt, R.G.; Rybczynski, R.; Lerner, M.R. Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: Comparisons with other insect OBPs and their signal peptides. J. Neurosci. 1991, 11, 2972–2984. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Prestwich, G.D. Expression and characterization of a lepidopteran general odorant binding protein. Insect Biochem. Mol. Biol. 1997, 27, 405–412. [Google Scholar] [CrossRef]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding protein-bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Plettner, E.; Lazar, J.; Prestwich, E.G.; Prestwich, G.D. Discrimination of pheromone enantiomers by two pheromone binding proteins from the Gypsy moth Lymantria dispar. Biochemistry 2000, 39, 8953–8962. [Google Scholar] [CrossRef]
- Terrado, M.; Pinnelli, G.R.; Sanes, J.; Plettner, E. Binding interactions, structure-activity relationships and blend effects in pheromone and host olfactory detection of herbivorous lepidoptera. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; Volume 2, Chapter 11; pp. 265–310. [Google Scholar]
- Zhou, J.J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori odorant-binding-proteins reveals that a general odorant-binding-protein discriminates between sex pheromone components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef] [Green Version]
- Hekmat-Scafe, D.S.; Steinbrecht, R.A.; Carlson, J.R. Coexpression of two odorant-binding protein homologs in Drosophila: Implications for olfactory coding. J. Neurosci. 1997, 17, 1616–1624. [Google Scholar] [CrossRef] [Green Version]
- Nardi, J.B.; Miller, L.A.; Walden, K.K.O.; Rovelstad, S.; Wang, L.P.; Frye, J.C.; Ramsdell, K.; Deem, L.S.; Robertson, H.M. Expression patterns of odorant-binding proteins in antennae of the moth Manduca sexta. Cell Tissue Res. 2003, 313, 321–333. [Google Scholar] [CrossRef]
- Maida, R.; Mameli, M.; Müller, B.; Krieger, J.; Steinbrecht, R.A. The expression pattern of four odorant-binding proteins in male and female silk moths, Bombyx mori. J. Neurocytol. 2005, 34, 149–163. [Google Scholar] [CrossRef]
- Shanbhag, S.R.; Smith, D.P.; Steinbrecht, R.A. Three odorant-binding proteins are co-expressed in sensilla trichodea of Drosophila melanogaster. Arthropod Struct. Dev. 2005, 34, 153–165. [Google Scholar] [CrossRef]
- Krieger, J.; von Nickisch-Roseneck, E.V.; Mameli, M.; Pelosi, P.; Breer, H. Binding proteins from the antennae of Bombyx mori. Insect Biochem. Mol. Biol. 1996, 26, 297–307. [Google Scholar] [CrossRef]
- Robertson, H.M.; Martos, R.; Sears, C.R.; Todres, E.Z.; Walden, K.K.O.; Nardi, J.B. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol. 1999, 8, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbach, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- Wojtasek, H.; Picimbon, J.F.; Leal, W.S. Identification and cloning of odorant binding proteins from the scarab beetle Phyllopertha diversa. Biochem. Biophys. Res. Commun. 1999, 263, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Biessmann, H.; Walter, M.F.; Dimitratos, S.; Woods, D. Isolation of cDNA clones from encoding putative odourant binding proteins from the antennae of the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol. Biol. 2002, 11, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F. Biochemistry and evolution of CSP and OBP proteins. In Insect Pheromone Biochemistry and Molecular Biology; Blomquist, G.J., Vogt, R.G., Eds.; Academic Press: New York, NY, USA, 2003; Chapter 11; pp. 539–566. [Google Scholar]
- McNeil, J.N.; Millar, J.G. Chemical communication: Pheromones and allelochemicals. In Insects: Structure and Function, 5th ed.; Chapman, R.F., Simpson, S.J., Douglas, A.E., Eds.; Cambridge University Press: Cambridge, UK, 2013; pp. 857–900. [Google Scholar]
- Story, R.N.; Keaster, A.J. Temporal and spatial distribution of black cutworms in midwest field crops. Environ. Entomol. 1982, 11, 1019–1022. [Google Scholar] [CrossRef]
- Showers, W.B.; Kaster, L.V.; Mulder, P.G. Corn seedling growth stage and black cutworm (Lepidoptera: Noctuidae) damage. Environ. Entomol. 1983, 12, 241–244. [Google Scholar] [CrossRef]
- Hong, S.C.; Williamson, R.C. Suitability of various turfgrass species and cultivars for development and survival of black cutworm (Lepidoptera: Noctuidae). J. Econ. Entomol. 2006, 99, 850–857. [Google Scholar] [CrossRef]
- Showers, W.B.; Keaster, A.J.; Raulston, J.R.; Hendrix, W.H., III; Derrick, M.E.; McCorcle, M.D.; Robinson, J.F.; Way, M.O.; Wallendorf, M.J.; Goodenough, J.L. Mechanism of southward migration of a noctuid moth [Agrotis ipsilon (Hufnagel)]: A complete migrant. Ecology 1993, 74, 2303–2314. [Google Scholar] [CrossRef]
- Sappington, T.W.; Showers, W.B. Implications for migration of age-related variation in flight behavior of Agrotis ipsilon (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 1991, 84, 560–565. [Google Scholar] [CrossRef]
- Picimbon, J.F.; Bécard, J.M.; Sreng, L.; Clément, J.L.; Gadenne, C. Juvenile hormone stimulates pheromonotropic brain factor release in the female black cutworm, Agrotis ipsilon. J. Insect Physiol. 1995, 41, 377–382. [Google Scholar] [CrossRef]
- Wynne, J.W.; Keaster, A.J.; Gerhardt, K.O.; Krause, G.F. Plant species identified as food sources for adult black cutworm (Lepidoptera:Noctuidae) in Northern Missouri. J. Kansas Entomol. Soc. 1991, 64, 381–387. [Google Scholar]
- Zhu, Y.; Keaster, A.J.; Gerhardt, K.O. Field observation on attractiveness of selected blooming plants to noctuid moths and electroantennogram responses of black cutworm (Lepidoptera: Noctuidae) moths to flower volatiles. Environ. Entomol. 1993, 22, 162–166. [Google Scholar] [CrossRef]
- Barrozo, R.B.; Jarriault, D.; Deisig, N.; Gemeno, C.; Monsempes, C.; Lucas, P.; Gadenne, C.; Anton, S. Mating-induced differential coding of plant-odour and sex pheromone in a male moth. Eur. J. Neurosci. 2011, 33, 1841–1850. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, W.H., III; Showers, W.B. Tracing black cutworm and armyworm (Lepidoptera: Noctuidae) northward migration using Pithecellobium and Calliandra pollen. Environ. Entomol. 1992, 21, 1092–1096. [Google Scholar] [CrossRef]
- Loublier, Y.; Douault, P.; Causse, R.; Barthes, J.; Buès, R.; Poitout, S.H. Utilisation des spectres polliniques recueillis sur Agrotis (Scotia) ipsilon hufnagel (Noctuidae) comme indicateur des migrations. Grana 1994, 33, 276–281. [Google Scholar] [CrossRef]
- Renou, M. Pheromones and general odor perception in insects. In Neurobiology of Chemical Communication; Mucignat-Caretta, C., Ed.; CRC Press: Boca Raton, FL, USA, 2014; Volume 1, pp. 23–56. [Google Scholar]
- Sell, C.S. Chemistry and the Sense of Smell; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; p. 480. [Google Scholar]
- Picimbon, J.F.; Gadenne, C.; Bécard, J.M.; Clément, J.L.; Sreng, L. Sex pheromone of the French black cutworm moth, Agrotis ipsilon (Lepidoptera, Noctuidae): Identification and regulation of a multicomponent blend. J. Chem. Ecol. 1997, 23, 211–230. [Google Scholar] [CrossRef]
- Gemeno, C.; Haynes, K. Chemical and behavioral evidence for a third pheromone component in a North American population of the black cutworm moth, Agrotis ipsilon. J. Chem. Ecol. 1998, 24, 999–1011. [Google Scholar] [CrossRef]
- Gemeno, C.; Lutfallah, A.F.; Haynes, K.F. Pheromone blend variation and cross-activation among populations of the black cutworm moth (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 2000, 93, 1322–1328. [Google Scholar] [CrossRef]
- Picimbon, J.F.; Gadenne, C. Evolution in noctuid pheromone binding proteins: Identification of PBP in the black cutworm moth, Agrotis ipsilon. Insect Biochem. Mol. Biol. 2002, 32, 839–846. [Google Scholar] [CrossRef]
- Abraham, D.; Löfstedt, C.; Picimbon, J.F. Molecular characterization and evolution of pheromone binding protein genes in Agrotis moths. Insect Biochem. Mol. Biol. 2005, 35, 1100–1111. [Google Scholar] [CrossRef]
- Gu, S.H.; Zhou, J.J.; Wang, G.R.; Zhang, Y.J.; Guo, Y.Y. Sex pheromone recognition and immunolocalization of three pheromone binding proteins in the black cutworm moth Agrotis ipsilon. Insect Biochem. Mol. Biol. 2013, 43, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Leal, W.S. Olfactory soluble proteins of cockroaches. Insect Biochem. Mol. Biol. 1999, 29, 973–978. [Google Scholar] [CrossRef]
- Walsh, M.J.; Mc Dougall, J.; Wittmann-Liebold, B. Extended N-terminal sequencing of proteins of archaebacterial ribosomes blotted from two-dimensional gels onto glass fiber and poly(vinylidene difluoride) membrane. Biochemistry 1988, 27, 6867–6876. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Asano, Y.; Ohtsuka, J.; Takada, Y.; Saito, K.; Ohki, R.; Fujii, H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci. Rep. 2013, 3, 3171. [Google Scholar] [CrossRef] [Green Version]
- Gadenne, C.; Picimbon, J.F.; Bécard, J.M.; Lalanne-Cassou, B.; Renou, M. Development and pheromone communication systems in hybrids of Agrotis ipsilon and Agrotis segetum (Lepidoptera: Noctuidae). J. Chem. Ecol. 1997, 23, 191–209. [Google Scholar] [CrossRef]
- Willett, C.S.; Harrison, R.G. Pheromone binding proteins in the European and Asian corn borers: No protein change associated with pheromone differences. Insect Biochem. Mol. Biol. 1999, 29, 277–284. [Google Scholar] [CrossRef]
- Ochieng, S.A.; Park, K.C.; Baker, T.C. Host-plant volatiles synergize responses of sex pheromone-specific olfactory receptors neurons in male Helicoverpa zea. J. Comp. Physiol. A 2002, 188, 325–333. [Google Scholar] [CrossRef]
- Pregitzer, P.; Schubert, M.; Breer, H.; Hansson, B.S.; Sachse, S.; Krieger, J. Plant odorants interfere with detection of sex pheromone signals by male Heliothis virescens. Front. Cell. Neurosci. 2012, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Rouyar, A.; Deisig, N.; Dupuy, F.; Limousin, D.; Wycke, M.A.; Renou, M.; Anton, S. Unexpected plant odor responses in a moth pheromone system. Front. Physiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouinguené, S.P.; Turlings, T.C.J. The effects of abiotic factors on induced volatile emissions in corn plants. Plant Physiol. 2002, 129, 1296–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñuelas, J.; Staudt, M. BVOCs and global change. Trends Plant Sci. 2010, 15, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Fares, S.; Park, J.H.; Gentner, D.R.; Weber, R.; Ormeño, E.; Karlik, J.; Goldstein, A.H. Seasonal cycles of biogenic volatile organic compounds fluxes and concentrations in a California citrus orchard. Atmos. Chem. Phys. 2012, 12, 9865–9880. [Google Scholar] [CrossRef] [Green Version]
- Najar-Rodriguez, A.; Schneeberger, M.; Bellutti, N.; Dorn, S. Variation in attraction to host plant odors in an invasive moth has a genetic basis and is genetically negatively correlated with fecundity. Behav. Genet. 2012, 42, 687–697. [Google Scholar] [CrossRef]
- Niinemets, Ü.; Kännaste, A.; Copolovici, L. Quantitative patterns between plant volatile emissions induced by biotic stresses and the degree of damage. Front. Plant Sci. 2013, 4, 262. [Google Scholar] [CrossRef] [Green Version]
- Blande, J.D.; Holopainen, J.K.; Niinemets, Ü. Plant volatiles in a polluted atmosphere: Stress response and signal degradation. Plant Cell Environ. 2014, 37, 1892–1904. [Google Scholar] [CrossRef] [Green Version]
- Loreto, F.; Dicke, M.; Schnitzler, J.P.; Turlings, T.C. Plant volatiles and the environment. Plant Cell Environ. 2014, 37, 1905–1908. [Google Scholar] [CrossRef]
- Degen, T.; Dillmann, C.; Marion-Poll, F.; Turlings, T.C.J. High genetic variability of herbivore-induced volatile emission within a broad range of maize inbred lines. Plant Physiol. 2004, 135, 1928–1938. [Google Scholar] [CrossRef] [Green Version]
- Vucetic, A.; Dahlin, I.; Petrovic-Obradovic, O.; Glinwood, R.; Webster, B.; Ninkovic, V. Volatile interaction between undamaged affects tritrophic interactions through changed plant volatile emission. Plant Signal. Behav. 2014, 9, e29517. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, P.; Krischik, V.A.; Jones, C.G. Microbial Mediation of Plant-Herbivore Interactions; John Wiley & Sons: New York, NY, USA, 1991; p. 530. [Google Scholar]
- Arguello, J.R.; Sellanes, C.; Lou, Y.R.; Raguso, R.A. Can yeast (S. cerevisiae) metabolic volatiles provide polymorphic signaling? PLoS ONE 2013, 8, e70219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turlings, T.C.J.; Loughrin, J.H.; McCall, P.J.; Röse, U.R.; Lewis, W.J.; Tumlinson, J.H. How caterpillar-damaged plants protect themselves by attracting parasitic wasps. Proc. Natl. Acad. Sci. USA 1995, 92, 4169–4174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turlings, T.C.J.; Tumlinson, J.H.; Lewis, W.J. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 1990, 250, 1251–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loughrin, J.H.; Manukian, A.; Heath, R.R.; Turlings, T.C.J.; Tumlinson, J.H. Diurnal cycle of emission of induced volatile terpenoids by herbivore-injured cotton plants. Proc. Natl. Acad. Sci. USA 1994, 91, 11836–11840. [Google Scholar] [CrossRef] [Green Version]
- De Moraes, C.M.; Lewis, W.J.; Paré, P.W.; Alborn, H.T.; Tumlinson, J.H. Herbivore-infested plants selectively attract parasitoids. Nature 1998, 393, 570–573. [Google Scholar] [CrossRef]
- Saxena, K.N.; Schoohoven, L.M. Induction of orientational and feeding preferences in Manduca sexta larvae for an artificial diet containing citral. Entomol. Exp. Appl. 1978, 23, 72–78. [Google Scholar] [CrossRef]
- Russell, C.; Wessnitzer, J.; Young, J.M.; Armstrong, J.D.; Webb, B. Dietary salt levels affect salt preference and learning in larval Drosophila. PLoS ONE 2011, 6, e20100. [Google Scholar] [CrossRef] [Green Version]
- Dormont, L.; Jay-Robert, P.; Bessière, J.M.; Rapior, S.; Lumaret, J.P. Innate olfactory preferences in dung beetles. J. Exp. Biol. 2010, 213, 3177–3186. [Google Scholar] [CrossRef] [Green Version]
- Sprayberry, J.D.H.; Ritter, K.A.; Riffell, J.A. The effect of olfactory exposure to non-insecticidal agrochemicals on bumblebee foraging behavior. PLoS ONE 2013, 8, e76273. [Google Scholar] [CrossRef] [Green Version]
- Kare, M.R.; Maller, O. The Chemical Senses and Nutrition; Elsevier Academic Press: New York, NY, USA, 2012; p. 510. [Google Scholar]
- Saveer, A.M.; Kromann, S.H.; Birgersson, G.; Bengtsson, M.; Lindblom, T.; Balkenius, A.; Hansson, B.S.; Witzgall, P.; Becher, P.G.; Ignell, R. Floral to green: Mating switches moth olfactory coding and preference. Proc. Biol. Sci. 2012, 279, 2314–2322. [Google Scholar] [CrossRef]
- Glinwood, R.; Ahmed, E.; Qvarfordt, E.; Ninkovic, V. Olfactory learning of plant genotypes by a polyphagous insect predator. Oecologia 2011, 166, 637–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todrank, J.; Heth, G.; Restrepo, D. Effects of in utero odorant exposure on neuroanatomical development of the olfactory bulb and odour preferences. Proc. Biol. Sci. 2011, 278, 1949–1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohl, J.V. Nutrient-dependent/pheromone-controlled adaptive evolution: A model. Socioaffective Neurosci. Psychol. 2013, 3, 20553. [Google Scholar] [CrossRef] [PubMed]
- Troemel, E.R.; Kimmel, B.E.; Bargmann, C.I. Reprogramming chemotaxis responses: Sensory neurons define olfactory preferences in C. elegans. Cell 1997, 91, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Glater, E.E.; Rockman, M.V.; Bargmann, C.I. Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens. G3 2014, 4, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Claudianos, C.; Lim, J.; Young, M.; Yan, S.; Cristino, A.S.; Newcomb, R.D.; Gunasekaran, N.; Reinhard, J. Odor memories regulate olfactory receptor expression in the sensory periphery. Eur. J. Neurosci. 2014, 39, 1642–1654. [Google Scholar] [CrossRef]
- Athrey, G.; Popkin-Hall, Z.; Veiga Cosme, L.; Takken, W.; Slotman, M.A. Species and sex-specific chemosensory gene expression in Anopheles coluzzii and An. quadriannulatus antennae. Parasit. Vectors 2020, 13, 212. [Google Scholar] [CrossRef]
- Riveron, J.; Boto, T.; Alcorta, E. Transcriptional basis of the acclimation to high environmental temperature at the olfactory receptor organs of Drosophila melanogaster. BMC Genom. 2013, 14, 259. [Google Scholar] [CrossRef] [Green Version]
- Dukes, J.P.; Deaville, R.; Bruford, M.W.; Youngson, A.F.; Jordan, W.C. Odorant receptor gene expression changes during the parr-smolt transformation in Atlantic salmons. Mol. Ecol. 2004, 13, 2851–2857. [Google Scholar] [CrossRef]
- Kamikouchi, A.; Morioka, M.; Kubo, T. Identification of honeybee antennal proteins/genes expressed in a sex- and/or caste selective manner. Zoolog. Sci. 2004, 21, 53–62. [Google Scholar] [CrossRef]
- Asgari, S.; Schmidt, O. Is cell surface calreticulin involved in phagocytosis by insect hemocytes? J. Insect Physiol. 2003, 49, 545–550. [Google Scholar] [CrossRef]
- Duan, Y.; Liu, P.; Li, J.; Wang, Y.; Li, J.; Chen, P. Molecular responses of calreticulin gene to Vibrio anguillarum and WSSV challenge in the ridgetail white prawn Exopalaemon carinicauda. Fish Shellfish Immunol. 2014, 36, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, J.R.; Horton, W.J.; Grotewiel, M.S. Odor-guided behavior in Drosophila requires calreticulin. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2003, 189, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M.; Corbett, E.F.; Mesaeli, N.; Nakamura, K.; Opas, M. Calreticulin: One protein, one gene, many functions. Biochem. J. 1999, 2, 281–292. [Google Scholar] [CrossRef]
- Deya, S.; Matsunamia, H. Calreticulin chaperones regulate functional expression of vomeronasal type 2 pheromone receptors. Proc. Natl. Acad. Sci. USA 2011, 108, 16651–16656. [Google Scholar] [CrossRef] [Green Version]
- Grotewiell, M.S.; Beckl, C.D.O.; Wu, K.H.; Zhu, X.R.; Davis, R.L. Integrin-mediated short-term memory in Drosophila. Nature 1998, 391, 455–460. [Google Scholar] [CrossRef]
- Xuan, N.; Guo, X.; Xie, H.Y.; Lou, Q.N.; Lu, X.B.; Liu, G.X.; Picimbon, J.F. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015, 22, 203–219. [Google Scholar] [CrossRef]
- Rabhi, K.K.; Esancy, K.; Voisin, A.; Crespin, L.; Le Corre, J.; Trichoire-Leignel, H.; Anton, S.; Gadenne, C. Unexpected effects of low doses of a neonicotinoid insecticide on behavioral responses to sex pheromone in a pest insect. PLoS ONE 2014, 9, e114411. [Google Scholar] [CrossRef] [Green Version]
- Einhorn, E.; Imler, J.L. Insect immunity: From systemic to chemosensory organs protection. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; Volume 2, Chapter 9; pp. 205–229. [Google Scholar]




| Protein Band | N-Terminal Sequence | Percent Identity | Clone Name | Species Source | Access Number |
|---|---|---|---|---|---|
| Aipsi1 | LTREEEANIKEAFHPFIMK | 90 | OBP4 | ipsilon | AGR39567 |
| 65 | OBP7 | armigera | AEB54591 | ||
| 65 | OBP7 | assulta | AGA16511 | ||
| 59 | ABP | virescens | CAC33574 | ||
| Aipsi2 | GVVMDEDMAELARMVRESCV | 100 | AipsABPX | ipsilon | AAP57463 |
| 89 | SinfABPX | inferens | AGS36754 | ||
| 89 | HvirABPX | virescens | CAA05508 | ||
| 89 | SexiABPX | exigua | AGP03461 | ||
| 85 | BmorABPX | mori | CAA64446 | ||
| 85 | MsexABPX | sexta | AAF16647 | ||
| Aipsi3 | TAEVMPHVTA | 90 | AipsGOBP2 | ipsilon | AAP57462 |
| 90 | AsegGOBP2 | segetum | ABI24161 | ||
| 90 | HvirGOBP2 | virescens | CAA65606 | ||
| 90 | BmorGOBP2 | mori | CAA64445 | ||
| 90 | MsexGOBP2 | sexta | AAG50015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picimbon, J.-F. Interpopulational Variations of Odorant-Binding Protein Expression in the Black Cutworm Moth, Agrotis ipsilon. Insects 2020, 11, 798. https://0-doi-org.brum.beds.ac.uk/10.3390/insects11110798
Picimbon J-F. Interpopulational Variations of Odorant-Binding Protein Expression in the Black Cutworm Moth, Agrotis ipsilon. Insects. 2020; 11(11):798. https://0-doi-org.brum.beds.ac.uk/10.3390/insects11110798
Chicago/Turabian StylePicimbon, Jean-François. 2020. "Interpopulational Variations of Odorant-Binding Protein Expression in the Black Cutworm Moth, Agrotis ipsilon" Insects 11, no. 11: 798. https://0-doi-org.brum.beds.ac.uk/10.3390/insects11110798