Genetic and Ecological Relationships of Anastrepha ludens (Diptera: Tephritidae) Populations in Southern Mexico
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Environmental Factors
2.2. Collection of Anastrepha Ludens Specimens and Host Plant Species
2.3. Genotyping
2.4. Population Genetic Analysis
2.4.1. Genetic Diversity, Male and Female Hardy–Weinberg Equilibrium Tests and Tests for Random Mating
2.4.2. Genetic Diversity by Factors Associated with Host Species and Locality
2.4.3. Genetic Structure
3. Results
3.1. Genetic Diversity of Males and Females per Locality
3.2. Genetic Diversity in Factors Associated with Host and Locality
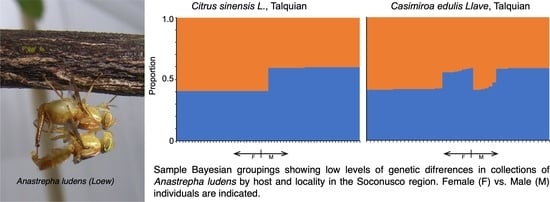
3.3. Genetic Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hartl, D.L.; Clark, A.G. Principles of Populations Genetics, 3rd ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 1997; pp. 118–119. [Google Scholar]
- Pinto Dias, N.; Zotti, M.J.; Montoya, P.; Carvalho, I.R.; Nava, D.E. Fruit fly management research: A systematic review of monitoring and control tactics in the world. Crop Prot. 2018, 112, 187–200. [Google Scholar] [CrossRef]
- Norrbom, A.L.; Kim, K.C. A List of Reported Host Plantas of the Species of Anastrepha Schiner; Plant Protection and Quarantine Program, U.S. Department of Agriculture, Animal and Plant Health Inspection Service: Riverdale, MD, USA, 1988; Volume 52, pp. 1–114. [Google Scholar]
- Hernández-Ortíz, V. El Género Anastrepha en México (Diptera-Tephritidae) Taxonomía, Distribución y Sus Plantas Hospederos; Instituto de Ecología, A.C., Sociedad Mexicana de Entomologia: Xalapa, México, 1992; p. 101. [Google Scholar]
- Aluja, M. Bionomics and managemente of Anastrepha. Annu. Rev. Entomol. 1994, 39, 155–178. [Google Scholar] [CrossRef]
- Dupuis, J.R.; Ruiz-Arce, R.; Barr, N.B.; Thomas, D.B.; Geib, S.M. Range-wide population genomics of the Mexican fruit fly: Toward development of pathway analysis tools. Evol. Appl. 2019, 12, 1641–1660. [Google Scholar] [CrossRef] [PubMed]
- Plummer, C.C.; McPhail, M.; Monk, J.W. The yellow chapote, a native host of the Mexican fruit fly. U.S. Dept. Agric. Tech. Bull. 1941, 775, 12. [Google Scholar]
- García Dessommes, G.J. El Origen de la Citricultura Moderna en México. In El Cultivo de Los Cítricos en el Estado de Nuevo León No. 1; Instituto Nacional de Investigaciones Frestales Agrícolas y Pecuarias: Ciudad de México, México; CIRNE, Campo Experimental General Terán: Terán, México, 2009; pp. 1–18. [Google Scholar]
- SIAP. Servicio de Información Agroalimentaria y Pesquera. Available online: http://infosiap.siap.gob.mx/images/stories/infogramas/100602-reporte-naranja.pdf. (accessed on 6 August 2020).
- Vanoye-Eligio, V.; Barrientos-Lozano, L.; Pérez-Castañeda, R.; Gaona-García, G.; Lara-Villalon, M. Population dynamics of Anastrepha ludens (Loew) (Diptera: Tephritidae) on citrus creas in Southern Tamaulipas, Mexico. Neotrop. Entomol. 2015, 44, 565–573. [Google Scholar] [CrossRef]
- Vanoye-Eligio, V.; Vázquez-Sauceda, M.L.; Rosas-Mejía, M.; Vera, A.; Cortés-Hernández, D.E.; Rocandio-Rodríguez, M. Analysis of temporal fluctuations in numbers of sexually mature Anastrepha ludens females over an extensive citrus area in northeastern Mexico. Entomol. Exp. Appl. 2019, 167, 517–525. [Google Scholar] [CrossRef]
- Futuyma, D.J.; Keese, M.C.; Funk, D.J. Genetic constraints on macroevolution: The evolution of host affiliation in the leaf beetle genus Ophraella. Evolution 1995, 49, 797–809. [Google Scholar] [CrossRef]
- Diehl, S.R.; Bush, G.L. An evolutionary and applied perspective of insect biotypes. Annu. Rev. Entomol. 1984, 29, 471–504. [Google Scholar] [CrossRef]
- Farrell, B.D.; Sequeira, A.S. Evolutionary rates in the adaptive radiaton of beetles on plants. Evolution 2004, 49, 1984–2001. [Google Scholar]
- Xie, X.; Rull, J.; Michel, A.P.; Velez, S.; Forbes, A.A.; Lobo, N.F.; Aluja, M.; Feder, J.L. Hawthorn-infesting populations of Rhagoletis pomonella in Mexico and speciation mode plurality. Evolution 2007, 61, 1091–1105. [Google Scholar] [CrossRef]
- Hood, G.R.; Powell, T.H.Q.; Doellman, M.M.; Sim, S.B.; Glover, M.; Yee, W.L.; Goughnour, R.B.; Mattsson, M.; Schwarz, D.; Feder, J.L. Rapid and repeatable host plant shifts drive reproductive isolation following a recent human-mediated introduction of the apple maggot fly, Rhagoletis pomonella. Evolution 2019, 74, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Feder, J.L.; Roethele, J.; Wlazlo, B.; Berlocher, S.H. Selective maintenance of allozyme differences among sympatric host races of the apple maggot fly. Proc. Natl. Acad. Sci. USA 1997, 94, 11417–11421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakovic, V.; Schuler, H.; Schebeck, M.; Feder, J.L.; Stauffer, C.; Ragland, G.J. Host plant-related genomic differentiation in the European cherry fruit fly, Rhagoletis cerasi. Mol. Ecol. 2019, 28, 4648–4666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Via, S. The ecological genetics of speciation. Am. Nat. 2002, 159, S1–S7. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Montoya, L.; Núñez-Farfán, J. Testing local host adaptation and phenotypic plasticity in a herbivore when alternative related host plants occur sympatrically. PLoS ONE 2013, 8, e79070. [Google Scholar] [CrossRef] [PubMed]
- Aluja, M.; Rull, J.; Pérez-Staples, D.; Díaz-Fleischer, F.; Sivinski, J. Random mating among Anastrepha ludens (Diptera: Tephritidae) adults of geographically distant and ecologically distinct populations in Mexico. Bull. Entomol. Res. 2009, 99, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Dávila-Jácome, A. Efecto del Cambio de Hospedero Larval Sobre los Parámetros Demográficos de la Mosca Mexicana de la Fruta Anastrepha Ludens (Loew) (Diptera: Tephritidae). Master’s Thesis, El Colegio de la Frontera Sur, Tapachula, México, 2004. [Google Scholar]
- Molina-Nery, M.C.; Ruiz-Montoya, L.; Zepeda-Cisneros, S.; Liedo, P. Genetic structure of populations of Anastrepha ludens (Diptera: Tephritidae) in Mexico. Fla. Entomol. 2014, 97, 1648–1661. [Google Scholar] [CrossRef]
- Ruiz-Arce, R.; Owen, C.L.; Thomas, D.B.; Barr, N.B.; McPheron, B.A. Phylogeographic structure in Anastrepha ludens (Diptera: Tephritidae) populations inferred with mtDNA sequencing. J. Econ. Entomol. 2015, 108, 1324–1336. [Google Scholar] [CrossRef] [Green Version]
- Pecina-Quintero, V.; Jiménez-Becerril, M.F.; Ruiz-Salazar, R.; Núñez-Colín, C.A.; Loera-Gallardo, J.L.; Hernández-Delgado, S.; Mayek-Pérez, N. Variability and genetic structure of Anastrepha ludens Loew (Diptera: Tephritidae) populations from Mexico. Int. J. Trop. Insect Sci. 2020, 40, 657–665. [Google Scholar] [CrossRef]
- Pecina-Quintero, V.; López Arroyo, J.; Loera Gallardo, J.; Rull, J.; Rosales Robles, E.; Cortez Mondaca, E.; Hernández Delgado, S.; Mayek Perez, N.; Aluja, M. Genetic differences between Anastrepha ludens Loew (Diptera: Tephritidae) populations stemming from a native and an exotic host in NE Mexico. Agric. Téc. Méx. 2009, 35, 323–331. [Google Scholar]
- Vera, M.T.; Cáceres, C.; Wornoayporn, V.; Islam, A.; Robinson, A.S.; de La Vega, M.H.; Hendrichs, J.; Cayol, J.P. Mating incompatibility among populations of the South American fruit fly Anastrepha fraterculus (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 2006, 99, 387–397. [Google Scholar] [CrossRef]
- Boykin, L.M.; Shatters, R.G.; Hall, D.G.; Burns, R.E.; Franqui, R.A. Analyisis of host preferences and geographical distribution of Anstrepha suspensa (Diptera Tephritidae) using phylogenetic analyses of mitchondrial cytochrome oxidase I DNA sequence data. Bull. Entomol. Res. 2006, 96, 457–469. [Google Scholar] [PubMed]
- Morgante, J.S.; Malavasi, A. Genetics variability in populations of the South American fruit fly Anastrepha fraterculus (Tephritidae). Rev. Bras. Gen. 1985, 8, 241–247. [Google Scholar]
- Hernández-Ortiz, V.; Bartolucci, A.F.; Morales-Valles, P.; Frías, D.; Selivon, D. Cryptic species of the Anastrepha fraterculus complex (Diptera: Tephritidae): A multivariate approach for the recognition of South American morphotypes. Ann. Entomol. Soc. Am. 2012, 105, 305–318. [Google Scholar] [CrossRef] [Green Version]
- Manni, M.; Lima, K.M.; Guglielmino, C.R.; Lanzavecchia, S.B.; Juri, M.; Vera, T.; Cladera, J.; Scolari, F.; Gomulski, L.; Bonizzoni, M.; et al. Relevant genetic differentiation among Brazilian populations of Anastrepha fraterculus (Diptera, tephritidae). ZooKeys 2015, 540, 157–173. [Google Scholar]
- Hedrick, P.W. Genetics of Populations, 2nd ed; Jones and Bartlett Publishers: Sudbury, MA, USA, 2000. [Google Scholar]
- Wilkinson, G.S.; Breden, F.; Mank, J.E.; Ritchie, M.G.; Higginson, A.D.; Radwan, J.; Jaquiery, J.; Salzburger, W.; Arriero, E.; Barriebeau, S.M.; et al. The locus of sexual selection: Moving sexual selection studies into the post-genomics era. J. Evol. Biol. 2015, 28, 739–755. [Google Scholar] [CrossRef] [Green Version]
- Mank, J.E.; Shu, J.J.; Wright, A.E. Signature of sexual conflict is actually conflict resolved. Mol. Ecol. 2020, 29, 215–217. [Google Scholar]
- Burk, T. Signaling and sex in acalyptrate flies. Fla. Entomol. 1981, 64, 30–43. [Google Scholar] [CrossRef]
- Gobierno del Estado de Chiapas. Programa Regional de Desarrollo 2013–2018; Gobierno del Estado de Chiapas: Tuxtla Gutiérrez, México, 2014; p. 75. [Google Scholar]
- Aluja, M.; Guillen, J.; De La Rosa, G.; Carrera, M.; Celedonio, H.; Liedo, P.; Hendrichs, J. Natural host plant survey of the economically important fruit flies (Diptera: Tephritidae) of Chiapas, Mexico. Fla. Entomol. 1987, 70, 329–338. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Beaton, M.J. Methodologies for Allozyme Analysis Using Cellulose Acetate Electrophoresis. Technical Manual of Cellulose Acetate Electrophoresis; Helena Laboratories: Beaumont, TX, USA, 1993; p. 32. [Google Scholar]
- Peakall, R. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. Biometrics, 2nd ed.; Freeman and Company: New York, NY, USA, 1995; p. 794. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puechmaille, S.J. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef] [PubMed]
- Celedonio-Hurtado, H.; Aluja, M.; Liedo, P. Population fluctuations of Anastrepha species (Diptera: Tephritidae) in tropical orchard habitats of Chiapas, Mexico. Environ. Entomol. 1995, 24, 861–869. [Google Scholar] [CrossRef]
- Behura, S.K. Molecular marker systems in insects: Current trends and future avenues. Mol. Ecol. 2006, 15, 3087–3113. [Google Scholar] [CrossRef]
- Aluja, M.; Mangan, R.L. Fruit Fly (Dipter: Tephritidae) host status determination: Critical conceptual, methodological, and regulatory considerations. Annu. Rev. Entomol. 2008, 53, 473–502. [Google Scholar] [CrossRef]
- Leal-Aguilar, K.; Ruiz-Montoya, L.; Perales, H.; Morales, H. Phenotypic plasticity of Brevicoryne brassicae in responses to nutritional quality of two related host plants. Ecol. Entomol. 2008, 33, 735–741. [Google Scholar]
- Chen, E.H.; Hou, Q.L.; Wei, D.D.; Jiang, H.B.; Wang, J.J. Phenotypic plasticity, trade-offs and gene expression changes accompanying dietary restriction and switches in Bactrocera dorsalis (Hendel) (Diptera: Tephritidae). Sci. Rep. 2017, 7, 1988. [Google Scholar] [CrossRef]
- Doellman, M.; Schuler, H.; Saint, G.J.; Glen, R.H.; Egan, S.P.; Powell, T.H.Q.; Glover, M.M.; Bruzzese, D.J.; Smith, J.J.; Yee, W.L.; et al. Geographic and ecological dimensions of host plant-associated genetic differentiation and speciation in the Rhagoletis cingulata (Diptera: Tephritidae) sibling species group. Insects 2019, 10, 275. [Google Scholar] [CrossRef] [Green Version]
- ECOSUR. Propuesta Interinstitucional Estatal de Ordenamiento Territorial; Laboratorio de Análisis de Información Geográfica y Estadística. El Colegio de la Frontera Sur: San Cristóbal de Las Casas, Chiapas, México, 2001; Available online: http://www.ecosur.mx/sitios/analisis-geografico/galeria/mapas-peot (accessed on 24 September 2020).
- Orozco-Dávila, D.; Hernández, R.; Meza, S.; Domínguez, J. Sexual competitiveness and compatibility between mass-reared sterile flies and wild populations of Anastrepha ludens (Diptera: Tephritidae) from different regions in Mexico. Fla. Entomol. 2007, 90, 19–26. [Google Scholar] [CrossRef]
- Fromhage, L.; Kokko, H.; Reid, J.M. Evolution of mate choice for genome-wide heterozygosity. Evolution 2009, 63, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Rowell-Rahier, M. Genetic structure of leaf-beetles populations: Microgeographic and sexual differentiation in Oreina cacaliae and O. speciosissima. Entomol. Exp. Appl. 1992, 65, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Foerster, K.; Delhey, K.; Johnsen, A.; Lifjel, T.J.; Kempanaers, B. Females increase offspring heterozygosity and fitness trhough extra-pair matings. Nature 2003, 425, 713–717. [Google Scholar] [CrossRef]
- Sánchez-Rosario, M.; Pérez-Staples, D.; Toledo, J.; Valle-Mora, J.; Liedo, P. Artificial selection on mating competitiveness of Anastrepha ludens for sterile insect technique application. Entomol. Exp. Appl. 2017, 162, 133–147. [Google Scholar] [CrossRef] [Green Version]
- Quintero-Fong, L.; Toledo, J.; Ruiz-Montoya, L.; Rendón, P.; Orozco-Dávila, D.; Valle-Mora, J.; Liedo, P. Demography of a genetic sexing strain of Anastrepha ludens (Diptera: Tephritidae): Effects of selection based on mating performance. Agric. Entomol. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Bosa, C.F.; Cruz-López, L.; Zepeda-Cisneros, C.S.; Valle-Mora, J.; Guillén-Navarro, K.; Liedo, P. Sexual behavior and male volatile compounds in wild and mass-reared strains of the Mexican fruit fly Anastrepha ludens (Diptera: Tephritidae) held under different colony management regimes. Insect Sci. 2014, 23, 105–116. [Google Scholar] [CrossRef]
- Teets, N.M.; Dias, V.S.; Pierce, B.K.; Schetelig, M.F.; Handler, A.M.; Hahn, D.A. Overexpression of an antioxidant enzyme improves male mating performance after stress in a lek-mating fruit fly. Proc. R. Soc. B Biol. Sci. 2019, 286, 29190531. [Google Scholar] [CrossRef] [Green Version]
- Rhainds, M. Ecology of female mating failure/lifelong virginity: A review of causal mechanisms in insects and arachnids. Entomol. Exp. Appl. 2019, 167, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.E.; Fumagalli, M.; Cooney, C.R.; Bloch, N.I.; Vieira, F.G.; Buechel, S.D.; Kolm, N.; Mank, J.E. Male-biased gene expression resolves sexual conflict through the evolution of sex-specific genetic architecture. Evol. Lett. 2018, 2, 52–61. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.E.; Rogers, T.F.; Fumagalli, M.; Cooney, C.R.; Mank, J.E. Phenotypic sexual dimorphism is associated wih genomic signature or resolved sexual conflict. Mol. Ecol. 2019, 28, 2860–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Doorn, G. Intralocus sexual conflict. Ann. N. Y. Acad. Sci. 2009, 1168, 52–71. [Google Scholar] [CrossRef] [PubMed]
- Courret, C.; Chang, C.-H.; Wei, K.H.C.; Montchamp-Moreau, C.; Larracuente, A.M. Sexually antagonistic selection promotes genetic divergence between males and females in an ant. Proc. R. Soc. B Biol. Sci. 2019, 116, 52–71. [Google Scholar]
- Harrison, R.G.; Bogdanowicz, S.M. Patterns of variation and linkage disequilibrium in field cricket hybrid zone. Evolution 1997, 51, 493–505. [Google Scholar] [CrossRef]
- Abdel Moniem, H.E.; Schemerhorn, B.J.; DeWoody, J.A.; Holland, J.D. Landscape genetics of a pollinator longhorn beetle [Typocerus v. velutinus (Olivier)] on a continuous habitat surface. Mol. Ecol. 2016, 25, 5015–5028. [Google Scholar] [CrossRef]
- Carey, J.R.; Harshman, L.G.; Liedo, P.; Müller, H.G.; Wang, J.L.; Zhang, Z. Longevity-fertility trade-offs in the tephritid fruit fly, Anastrepha ludens, across dietary-restriction gradients. Aging Cell 2008, 7, 470–477. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.B.; Loera-Gallardo, J. Dispersal and longevity of mass-released, sterilized Mexican fruit flies (Diptera: Tephritidae). Environ. Entomol. 1998, 27, 1045–1052. [Google Scholar] [CrossRef]
- Arnold, S.J.; Wade, M.J. On the measurement of natural and sexual selection: Applications. Evolution 1984, 38, 720–734. [Google Scholar] [CrossRef]
- Darwin, C. The Descent of Man, and Selection in Relation to Sex; Johen Murray: London, UK, 1871; p. 254. [Google Scholar]
- Alberti, A.C.; Calcagno, G.; Saidman, B.O.; Vilardi, J.C. Analysis of the genetic structure of a natural population of Anastrepha fraterculus (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 1999, 92, 731–736. [Google Scholar] [CrossRef]
- McInnis, D.O.; Lance, D.R.; Jackson, C.G. Behavioral resistance to the sterile insect technique by Mediterranean fruit fly (Diptera: Tephritidae) in Hawaii. Ann. Entomol. Soc. Am. 1996, 89, 739–744. [Google Scholar] [CrossRef]



| Locality Name | Longitude | Latitude | Elevation (m) | CC | CP | MI | CE |
|---|---|---|---|---|---|---|---|
| Reforma | −92.32851614 | 15.0403897 | 387 | x | |||
| Guadalupe | −92.27440112 | 15.01203335 | 427 | x | |||
| El Triunfo | −92.22400912 | 14.98588244 | 470 | x | x | ||
| San Carlos | −92.26178333 | 15.04050278 | 532 | x | |||
| Toluca | −92.24566958 | 15.02878192 | 538 | x | |||
| Salvador Urbina | −92.21133333 | 15.03722222 | 540 | x | |||
| El Eden | −92.30475556 | 15.05781389 | 551 | x | x | ||
| Ahuacatlan | −92.17528333 | 15.042325 | 719 | x | |||
| Santo Domingo | −92.09836944 | 15.03047222 | 918 | x | |||
| Unión Juárez | −92.07942076 | 15.06620901 | 1386 | x | |||
| Talquian | −92.08379166 | 15.08690091 | 1696 | x | x |
| Locality (Host) | Sex | N | Na | Ho | He | f | P |
|---|---|---|---|---|---|---|---|
| Reforma (Mi) | Female | 20 | 2.3 | 0.317 | 0.451 | 0.245 | 100.00 |
| Male | 20 | 2.0 | 0.467 | 0.434 | −0.091 | 100.00 | |
| 2.5 | 0.392 | 0.528 | 0.241 | 100.00 | |||
| Guadalupe (Cp) | Female | 30 | 1.8 | 0.056 | 0.215 | 0.812 | 66.67 |
| Male | 30 | 2.3 | 0.178 | 0.468 | 0.630 | 100.00 | |
| 2.3 | 0.117 | 0.410 | 0.731 | 100.00 | |||
| El Triunfo (Ce, Mi) | Female | 40 | 2.2 | 0.188 | 0.360 | 0.452 | 83.33 |
| Male | 40 | 2.3 | 0.329 | 0.517 | 0.362 | 100.00 | |
| 2.3 | 0.258 | 0.517 | 0.503 | 100.00 | |||
| San Carlos (Cc) | Female | 29 | 2.2 | 0.241 | 0.369 | 0.490 | 83.33 |
| Male | 30 | 2.0 | 0.189 | 0.299 | 0.538 | 83.33 | |
| 2.3 | 0.215 | 0.447 | 0.646 | 100.00 | |||
| Toluca (Cc) | Female | 30 | 2.0 | 0.028 | 0.410 | 0.933 | 83.33 |
| Male | 30 | 1.2 | 0.000 | 0.083 | 1.000 | 16.67 | |
| 2.2 | 0.014 | 0.379 | 0.961 | 100.00 | |||
| Salvador Urbina (Cc) | Female | 30 | 2.0 | 0.089 | 0.354 | 0.671 | 83.33 |
| Male | 30 | 1.3 | 0.006 | 0.148 | 0.963 | 33.33 | |
| 2.5 | 0.392 | 0.528 | 0.241 | 83.33 | |||
| Edén (Cc) | Female | 42 | 2.5 | 0.190 | 0.445 | 0.493 | 100.00 |
| Male | 49 | 2.2 | 0.197 | 0.413 | 0.543 | 83.33 | |
| 2.5 | 0.194 | 0.484 | 0.611 | 100.00 | |||
| Ahuacatlán (Cc) | Female | 27 | 2.0 | 0.154 | 0.241 | 0.610 | 66.67 |
| Male | 20 | 1.8 | 0.250 | 0.341 | 0.302 | 66.67 | |
| 2.2 | 0.195 | 0.334 | 0.566 | 83.33 | |||
| Santo Domingo (Cc) | Female | 30 | 2.3 | 0.117 | 0.391 | 0.710 | 100.00 |
| Male | 30 | 2.0 | 0.222 | 0.368 | 0.477 | 83.33 | |
| 2.3 | 0.169 | 0.417 | 0.637 | 100.00 | |||
| Unión Juárez (Cc) | Female | 31 | 2.2 | 0.102 | 0.247 | 0.643 | 83.33 |
| Male | 30 | 1.8 | 0.106 | 0.249 | 0.481 | 66.67 | |
| 2.3 | 0.104 | 0.339 | 0.725 | 100.00 | |||
| Talquian (Cc, Ce) | Female | 58 | 2.2 | 0.075 | 0.412 | 0.823 | 100.00 |
| Male | 50 | 2.0 | 0.113 | 0.331 | 0.658 | 83.33 | |
| 2.3 | 0.093 | 0.488 | 0.798 | 100.00 | |||
| Grand mean over loci and populations (22) | Mean | 33 | 2.0 | 0.337 | 0.343 | 0.545 | 80.30 |
| SE | 0.0 | 0.018 | 0.018 | 0.038 | 4.61 |
| Locality | Females | Males | ||||
|---|---|---|---|---|---|---|
| χ2 | p | d.f. | χ2 | p | d.f. | |
| Reforma | 38.4 | 0.0001 | 12 | 25.7 | 0.0120 | 12 |
| Guadalupe | 117.6 | <0.0001 | 10 | 101.0 | 0.0000 | 12 |
| El Triunfo | 103.9 | <0.0001 | 10 | 59.0 | <0.0001 | 12 |
| San Carlos | 99.7 | <0.0001 | 8 | 129.7 | <0.0001 | 10 |
| Toluca | 149.0 | <0.0001 | 10 | 30.0 | <0.0001 | 1 |
| Salvador Urbina | 124.3 | <0.0001 | 10 | 19.1 | 0.0007 | 4 |
| Edén | 128.8 | <0.0001 | 8 | 109.9 | <0.0001 | 5 |
| Ahuacatlán | 121.1 | <0.0001 | 8 | 50.1 | <0.0001 | 8 |
| Santo Domingo | 128.8 | <0.0001 | 12 | 89.9 | <0.0001 | 10 |
| Unión Juárez | 105.1 | <0.0001 | 8 | 40.1 | <0.0001 | 10 |
| Talquian | 276.9 | <0.0001 | 12 | 134.5 | <0.0001 | 10 |
| Locality | Locus | |||||
|---|---|---|---|---|---|---|
| 6PGDH a | G6PDH b | GOT a | IDH b | ME a | PGM b | |
| Reforma | 3.96 NS | 30.37 *** | 10.97 * | 30.04 *** | 16.77 *** | 28.34 *** |
| Guadalupe | 20.18 *** | 268.97 *** | 44.90 *** | 46.24 *** | 32.49 *** | 56.30 *** |
| El Triunfo | 76.51 *** | 88.20 *** | 50.69 *** | 59.01 *** | 9.90 *** | 472.38 *** |
| San Carlos | 52.60 *** | 30.85 *** | 38.19 *** | 24.50 *** | 859.43 *** | 39.77 *** |
| Toluca | 103.56 *** | 1296.00 *** | 59.84 *** | 233.68 *** | M | 317.04 *** |
| Salvador Urbina | 344.20 *** | 2649.05 *** | 50.44 *** | 76.92 *** | M | 126.56 *** |
| Edén | 27.02 *** | 61.30 *** | 60.67 *** | 44.04 *** | 64.60 *** | 287.42 *** |
| Ahuacatlán | 25.30 *** | 15.91 *** | 39.49 *** | 34.05 *** | 161.87 *** | M |
| Santo Domingo | 29.05 *** | 22.40 *** | 55.19 *** | 16.84 ** | 80.97 *** | 50.32 *** |
| Unión Juárez | 1.00 NS | 9.38 * | 44.11 *** | 69.07 *** | 595.12 *** | 147.80 *** |
| Talquian | 268.75 *** | 4071.77 *** | 89.56 *** | 44.28 *** | 97.75 *** | 671.46 *** |
| Host | Locality | N | P | Na | Ho | He | f | Mean | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | N | Na | Ho | He | f | ||||||||
| Citrus sinensis | San Carlos | 59 | 100.0 | 2.333 | 0.215 | 0.447 | 0.646 | 91.67 | 56 | 2.271 | 0.118 | 0.397 | 0.744 |
| Toluca | 60 | 83.33 | 2.167 | 0.014 | 0.379 | 0.961 | |||||||
| Salvador Urbina | 60 | 83.33 | 2.167 | 0.047 | 0.381 | 0.896 | |||||||
| Edén | 44 | 83.33 | 2.167 | 0.110 | 0.409 | 0.740 | |||||||
| Ahuacatlán | 47 | 83.33 | 2.333 | 0.191 | 0.337 | 0.580 | |||||||
| Santo Domingo | 60 | 100.0 | 2.333 | 0.169 | 0.417 | 0.637 | |||||||
| Unión Juárez | 61 | 100.0 | 2.333 | 0.104 | 0.339 | 0.725 | |||||||
| Talquián | 60 | 100.0 | 2.333 | 0.094 | 0.470 | 0.769 | |||||||
| Casimiroa edulis | El Triunfo | 40 | 100.0 | 2.167 | 0.171 | 0.447 | 0.599 | 91.67 | 49 | 2.167 | 0.126 | 0.414 | 0.697 |
| Talquián | 48 | 83.33 | 2.000 | 0.090 | 0.386 | 0.761 | |||||||
| Citrus paradisi | Guadalupe | 60 | 100.0 | 2.333 | 0.117 | 0.410 | 0.731 | ||||||
| Mangifera indica | Reforma | 40 | 100.0 | 2.500 | 0.392 | 0.528 | 0.241 | 100.0 | 42 | 2.444 | 0.337 | 0.497 | 0.911 |
| El Triunfo | 40 | 100.0 | 2.333 | 0.346 | 0.491 | 0.296 | |||||||
| Edén | 47 | 100.0 | 2.500 | 0.273 | 0.474 | 0.366 | |||||||
| Mean over 14 samples | 94.05 | 52 | 2.286 | 0.167 | 0.422 | 0.301 | |||||||
| Hierarchical Arrangement | Source of Variation | d.f. | Sum of Squares | Medium Square | Estimated Variance | Variance Component (%) | Differentiation Estimation |
|---|---|---|---|---|---|---|---|
| Locality, sex | Among localities | 10 | 482.93 | 48.29 | 0.00 | 0 | Φ loc = 0.0 Φ sex(loc) = 0.386 * Φ (total) = 0.243 * |
| Between sexes (locality) | 11 | 784.20 | 71.29 | 2.067 | 39 | ||
| Within individuals | 704 | 2318.50 | 3.29 | 3.29 | 61 | ||
| Total | 725 | 3585.63 | 5.4 | 100 | |||
| Localities | Among localities | 14 | 653.902 | 50.3 | 0.892 | 18 | Φ = 0.178 *** |
| Within individuals | 712 | 2931.73 | 4.12 | 4.12 | 82 | ||
| Total | 725 | 3585.63 | 5.01 | 100 | |||
| Hosts | Among hosts | 3 | 292.4 | 97.47 | 0.683 | 13 | Φ = 0.13 *** |
| Within individuals | 722 | 3293.2 | 4.56 | 4.561 | 87 | ||
| Total | 725 | 3585.6 | 5.244 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Montoya, L.; Vallejo, R.V.; Haymer, D.; Liedo, P. Genetic and Ecological Relationships of Anastrepha ludens (Diptera: Tephritidae) Populations in Southern Mexico. Insects 2020, 11, 815. https://0-doi-org.brum.beds.ac.uk/10.3390/insects11110815
Ruiz-Montoya L, Vallejo RV, Haymer D, Liedo P. Genetic and Ecological Relationships of Anastrepha ludens (Diptera: Tephritidae) Populations in Southern Mexico. Insects. 2020; 11(11):815. https://0-doi-org.brum.beds.ac.uk/10.3390/insects11110815
Chicago/Turabian StyleRuiz-Montoya, Lorena, Rodrigo Verónica Vallejo, David Haymer, and Pablo Liedo. 2020. "Genetic and Ecological Relationships of Anastrepha ludens (Diptera: Tephritidae) Populations in Southern Mexico" Insects 11, no. 11: 815. https://0-doi-org.brum.beds.ac.uk/10.3390/insects11110815