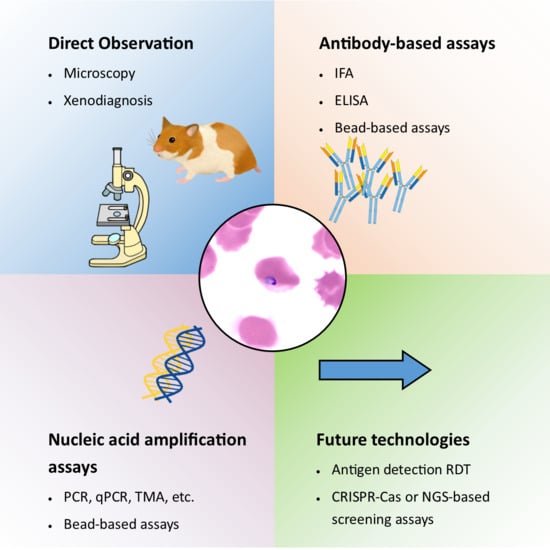
Technologies for Detection of Babesia microti: Advances and Challenges
Abstract
:1. Introduction
2. Babesia microti Biology and Detection Challenges
3. Detection Techniques
3.1. Direct Demonstration of B. microti Parasites
3.1.1. Experimental Inoculation
3.1.2. Blood Film Microscopy
3.2. Detection of Biomarkers of B. microti Infection
3.2.1. Nucleic Acid-Based Assays
3.2.2. Antigen Detection Assays
3.2.3. Antibody-Based Assays
4. Multiplex Assays
5. Novel and Future Technologies
5.1. Next Generation Sequencing
5.2. CRISPR Technology
6. Blood Donor Screening
7. Assay Validation
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Homer, M.J.; Aguilar-Delfin, I.; Telford, S.R., 3rd; Krause, P.J.; Persing, D.H. Babesiosis. Clin. Microbiol. Rev. 2000, 13, 451–469. [Google Scholar] [CrossRef]
- Hunfeld, K.-P.; Hildebrandt, A.; Gray, J.S. Babesiosis: Recent insights into an ancient disease. Int. J. Parasitol. 2008, 38, 1219–1237. [Google Scholar] [CrossRef]
- Vannier, E.; Krause, P.J. Human babesiosis. N. Engl. J. Med. 2012, 366, 2397–2407. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Hong-Xiang, Z.; Huang, J.-L.; Tambo, E.; Zhuge, H.-X.; Zhou, X.-N. Human babesiosis, an emerging tick-borne disease in the People’s Republic of China. Parasites Vectors 2014, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Babes, V. Sur l’hemoglobinuria bacterienne des boeufs. Comptes Rendus De L’académie Des Sci. 1888, 107, 693–694. [Google Scholar]
- Skrabalo, Z.; Deanovic, Z. Piroplasmosis in man; report of a case. Doc. Med. Geogr. Trop. 1957, 9, 11–16. [Google Scholar] [PubMed]
- Telford, S.R.; Gorenflot, A.; Brasseur, P.; Spielman, A. Babesial infections in humans and wildlife. In Parasitic Protozoa, 2nd ed.; Kreier, J.P., Ed.; Academic Press: San Diego, CA, USA, 1993; pp. 1–47. [Google Scholar]
- Scholtens, R.G.; Gleason, N.; Braff, E.H.; Healy, G.R. A case of babesiosis in man in the United States. Am. J. Trop. Med. Hyg. 1968, 17, 810–813. [Google Scholar] [CrossRef]
- Piesman, J.; Spielman, A. Human babesiosis on Nantucket Island: Prevalence of Babesia microti in ticks. Am. J. Trop. Med. Hyg. 1980, 29, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.B.; Herwaldt, B.L. Babesiosis surveillance—United States, 2011–2015. MMWR Surveill. Summ. 2019, 68, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Western, K.A.; Benson, G.D.; Gleason, N.N.; Healy, G.R.; Schultz, M.G. Babesiosis in a Massachusetts Resident. N. Engl. J. Med. 1970, 283, 854–856. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Surveillance for Babesiosis—United States, 2019 Annual Summary. 2021. Available online: https://www.cdc.gov/parasites/babesiosis/resources/Surveillance_Babesiosis_US_2019.pdf. (accessed on 23 November 2021).
- Vannier, E.G.; Diuk-Wasser, M.A.; Ben Mamoun, C.; Krause, P.J. Babesiosis. Infect. Dis. Clin. North Am. 2015, 29, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Vannier, E.; Krause, P.J. 100—Babesiosis. In Hunter’s Tropical Medicine and Emerging Infectious Disease (Ninth Edition); Magill, A.J., Hill, D.R., Solomon, T., Ryan, E.T., Eds.; W.B. Saunders: London, UK, 2013; pp. 761–763. [Google Scholar]
- Ruebush, T.K., 2nd; Collins, W.E.; Healy, G.R.; Warren, M. Experimental Babesia microti infections in non-splenectomized Macaca mulatta. J. Parasitol. 1979, 65, 144–146. [Google Scholar] [PubMed]
- Ii, T.K.R.; Warren, M.; Spielman, A.; Collins, W.E.; Piesman, J. Tick Transmission of Babesia microti to Rhesus Monkeys (Macaca mulatta). Am. J. Trop. Med. Hyg. 1981, 30, 555–559. [Google Scholar] [CrossRef]
- White, D.J.; Talarico, J.; Chang, H.-G.; Birkhead, G.S.; Heimberger, T.; Morse, D.L. Human babesiosis in New York State: Review of 139 hospitalized cases and analysis of prognostic factors. Arch. Intern. Med. 1998, 158, 2149–2154. [Google Scholar] [CrossRef]
- Hatcher, J.C.; Greenberg, P.D.; Antique, J.; Jimenez-Lucho, V.E. Severe babesiosis in Long Island: Review of 34 cases and their complications. Clin. Infect. Dis. 2001, 32, 1117–1125. [Google Scholar] [CrossRef]
- Sun, T.; Tenenbaum, M.J.; Greenspan, J.; Teichberg, S.; Wang, R.T.; Degnan, T.; Kaplan, M.H. Morphologic and clinical observations in human infection with Babesia microti. J. Infect. Dis. 1983, 148, 239–248. [Google Scholar] [CrossRef]
- Krause, P.J.; McKay, K.; Gadbaw, J.; Christianson, D.; Closter, L.; Lepore, T.; Telford, S.R., 3rd; Sikand, V.; Ryan, R.; Persing, D.; et al. Increasing health burden of human babesiosis in endemic sites. Am. J. Trop. Med. Hyg. 2003, 68, 431–436. [Google Scholar] [CrossRef]
- Krause, P.J.; Spielman, A.; Telford, S.R., 3rd; Sikand, V.K.; McKay, K.; Christianson, D.; Pollack, R.J.; Brassard, P.; Magera, J.; Ryan, R.; et al. Persistent parasitemia after acute babesiosis. N. Engl. J. Med. 1998, 339, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Moritz, E.D.; Winton, C.S.; Tonnetti, L.; Townsend, R.L.; Berardi, V.P.; Hewins, M.-E.; Weeks, K.E.; Dodd, R.Y.; Stramer, S.L. Screening for Babesia microti in the U.S. blood supply. N. Engl. J. Med. 2016, 375, 2236–2245. [Google Scholar] [CrossRef] [Green Version]
- Gorenflot, A.; Moubri, K.; Precigout, E.; Carcy, B.; Schetters, T.P. Human babesiosis. Ann. Trop. Med. Parasitol. 1998, 92, 489–501. [Google Scholar] [CrossRef]
- Krause, P.J.; Telford, S., 3rd; Spielman, A.; Ryan, R.; Magera, J.; Rajan, T.V.; Christianson, D.; Alberghini, T.V.; Bow, L.; Persing, D. Comparison of PCR with blood smear and inoculation of small animals for diagnosis of Babesia microti parasitemia. J. Clin. Microbiol. 1996, 34, 2791–2794. [Google Scholar] [CrossRef] [Green Version]
- Krause, P.J.; Telford, S.R., 3rd; Ryan, R.; Conrad, P.A.; Wilson, M.; Thomford, J.W.; Spielman, A. Diagnosis of babesiosis: Evaluation of a serologic test for the detection of Babesia microti antibody. J. Infect. Dis. 1994, 169, 923. [Google Scholar] [CrossRef]
- Laveran, A. Un nouveau parasite trouvé dans le sang des malades atteints de fièvre palustre: Origine parasitaire des accidents de l’impaludisme; J.-B. Baillière: Paris, France, 1881. [Google Scholar]
- Healy, G.R.; Ruebush, T.K., 2nd. Morphology of Babesia microti in human blood smears. Am. J. Clin. Pathol. 1980, 73, 107–109. [Google Scholar] [CrossRef] [Green Version]
- Trampuz, A.; Jereb, M.; Muzlovic, I.; Prabhu, R.M. Clinical review: Severe malaria. Crit. Care 2003, 7, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Wongsrichanalai, C.; Barcus, M.J.; Muth, S.; Sutamihardja, A.; Wernsdorfer, W.H. A review of malaria diagnostic tools: Microscopy and rapid diagnostic test (RDT). Am. J. Trop. Med. Hyg. 2007, 77, 119–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, D.; Wongsrichanalai, C.; Barnwell, J.W. Ensuring quality and access for malaria diagnosis: How can it be achieved? Nat. Rev. Microbiol. 2006, 4, S7–S20. [Google Scholar] [CrossRef] [PubMed]
- Vannier, E.; Gewurz, B.E.; Krause, P.J. Human babesiosis. Infect. Dis. Clin. North Am. 2008, 22, 469–488. [Google Scholar] [CrossRef] [Green Version]
- Persing, D.H. PCR detection of Babesia microti. In Diagnostic Molecular Microbiology: Principles and Applications; Persing, D.H., Tenover, F.C., White, T.J., Eds.; American Society for Microbiology: Washington, DC, USA, 1993; pp. 475–479. [Google Scholar]
- Persing, D.H.; Mathiesen, D.; Marshall, W.F.; Telford, S.R.; Spielman, A.; Thomford, J.W.; Conrad, P.A. Detection of Babesia microti by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 2097–2103. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Villafuerte, P.; Zhuge, J.; Visintainer, P.; Wormser, G.P. Comparison of a quantitative PCR assay with peripheral blood smear examination for detection and quantitation of Babesia microti infection in humans. Diagn. Microbiol. Infect. Dis. 2015, 82, 109–113. [Google Scholar] [CrossRef]
- Tonnetti, L.; Young, C.; Kessler, D.A.; Williamson, P.C.; Reik, R.; Proctor, M.C.; Brès, V.; Deisting, B.; Bakkour, S.; Schneider, W.; et al. Transcription-mediated amplification blood donation screening for Babesia. Transfusion 2020, 60, 317–325. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization & UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases. Microscopy for the Detection, Identification and Quantification of Malaria Parasites on Stained Thick and Thin Blood Films in Research Settings (Version 1.0): Procedure: Methods Manual. World Health Organization. 2015. Available online: https://apps.who.int/iris/handle/10665/163782 (accessed on 14 November 2021).
- Sethabutr, O.; Brown, A.E.; Panyim, S.; Kain, K.C.; Webster, H.K.; Echeverria, P. Detection of plasmodium falciparum by polymerase chain reaction in a field study. J. Infect. Dis. 1992, 166, 145–148. [Google Scholar] [CrossRef]
- Hoare, C.A. Comparative aspects of human babesiosis. Trans. R. Soc. Trop. Med. Hyg. 1980, 74, 143–148. [Google Scholar] [CrossRef]
- Wang, G.; Wormser, G.P.; Zhuge, J.; Villafuerte, P.; Ip, D.; Zeren, C.; Fallon, J.T. Utilization of a real-time PCR assay for diagnosis of Babesia microti infection in clinical practice. Ticks Tick Borne Dis. 2015, 6, 376–382. [Google Scholar] [CrossRef]
- Bloch, E.M.; Lee, T.-H.; Krause, P.J.; Telford, S.R., 3rd; Montalvo, L.; Chafets, D.; Usmani-Brown, S.; Lepore, T.J.; Busch, M.P. Development of a real-time polymerase chain reaction assay for sensitive detection and quantitation of Babesia microti infection. Transfusion 2013, 53, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.; Glaser, K.C.; Adams-Fish, D.; Boley, M.; Mayda, M.; Molestina, R.E. Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Exp. Parasitol. 2015, 149, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Cornillot, E.; Hadj-Kaddour, K.; Dassouli, A.; Noel, B.; Ranwez, V.; Vacherie, B.; Augagneur, Y.; Bres, V.; Duclos, A.; Randazzo, S.; et al. Sequencing of the smallest apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 2012, 40, 9102–9114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.C.; Cornillot, E.; McCracken, C.; Usmani-Brown, S.; Dwivedi, A.; Ifeonu, O.O.; Crabtree, J.; Gotia, H.T.; Virji, A.Z.; Reynes, C.; et al. Genome-wide diversity and gene expression profiling of Babesia microti isolates identify polymorphic genes that mediate host-pathogen interactions. Sci. Rep. 2016, 6, 35284. [Google Scholar] [CrossRef]
- Verma, N.; Puri, A.; Essuman, E.; Skelton, R.; Anantharaman, V.; Zheng, H.; White, S.; Gunalan, K.; Takeda, K.; Bajpai, S.; et al. Antigen discovery, bioinformatics and biological characterization of novel immunodominant Babesia microti antigens. Sci. Rep. 2020, 10, 9598. [Google Scholar] [CrossRef] [PubMed]
- Grabias, B.; Clement, J.; Krause, P.J.; Lepore, T.; Kumar, S. Superior real-time polymerase chain reaction detection of Babesia microti parasites in whole blood utilizing high-copy BMN antigens as amplification targets. Transfusion 2018, 58, 1924–1932. [Google Scholar] [CrossRef]
- Bakkour, S.; Chafets, D.M.; Wen, L.; Muench, M.; Telford, S.R., 3rd; Erwin, J.L.; Levin, A.E.; Self, D.; Brès, V.; Linnen, J.M.; et al. Minimal infectious dose and dynamics of Babesia microti parasitemia in a murine model. Transfusion 2018, 58, 2903–2910. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, G.L.; Edlind, T.D.; Ihler, G.M. Detection of Babesia bovis using DNA hybridization1. J. Protozool. 1986, 33, 125–128. [Google Scholar] [CrossRef]
- Lemieux, J.E.; Tran, A.D.; Freimark, L.; Schaffner, S.F.; Goethert, H.; Andersen, K.G.; Bazner, S.; Lisa, F.; McGrath, G.; Sloan, L.; et al. A global map of genetic diversity in Babesia microti reveals strong population structure and identifies variants associated with clinical relapse. Nat. Microbiol. 2016, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Baniecki, M.L.; Moon, J.; Sani, K.; Lemieux, J.E.; Schaffner, S.F.; Sabeti, P.C. Development of a SNP barcode to genotype Babesia microti infections. PLOS Negl. Trop. Dis. 2019, 13, e0007194. [Google Scholar] [CrossRef]
- Lodes, M.J.; Houghton, R.L.; Bruinsma, E.S.; Mohamath, R.; Reynolds, L.D.; Benson, D.R.; Krause, P.J.; Reed, S.G.; Persing, D.H. Serological expression cloning of novel immunoreactive antigens of Babesia microti. Infect. Immun. 2000, 68, 2783–2790. [Google Scholar] [CrossRef] [Green Version]
- Homer, M.J.; Lodes, M.J.; Reynolds, L.D.; Zhang, Y.; Douglass, J.F.; McNeill, P.D.; Houghton, R.L.; Persing, D.H. Identification and characterization of putative secreted antigens from Babesia microti. J. Clin. Microbiol. 2003, 41, 723–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornillot, E.; Dassouli, A.; Pachikara, N.; Lawres, L.; Renard, I.; Francois, C.; Randazzo, S.; Brès, V.; Garg, A.; Brancato, J.; et al. A targeted immunomic approach identifies diagnostic antigens in the human pathogen Babesia microti. Transfusion 2016, 56, 2085–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Jia, H.; Terkawi, M.A.; Goo, Y.-K.; Kawano, S.; Ooka, H.; Li, Y.; Yu, L.; Cao, S.; Yamagishi, J.; et al. Identification and characterization of a novel secreted antigen 1 of Babesia microti and evaluation of its potential use in enzyme-linked immunosorbent assay and immunochromatographic test. Parasitol. Int. 2011, 60, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Thekkiniath, J.; Mootien, S.; Lawres, L.; Perrin, B.A.; Gewirtz, M.; Krause, P.J.; Williams, S.; Doggett, J.S.; Ledizet, M.; Ben Mamoun, C. BmGPAC, an antigen capture assay for detection of active Babesia microti infection. J. Clin. Microbiol. 2018, 56, e00067-18. [Google Scholar] [CrossRef] [Green Version]
- Homer, M.J.; Bruinsma, E.S.; Lodes, M.J.; Moro, M.H.; Telford, S., 3rd; Krause, P.J.; Reynolds, L.D.; Mohamath, R.; Benson, D.R.; Houghton, R.L.; et al. A polymorphic multigene family encoding an immunodominant protein from Babesia microti. J. Clin. Microbiol. 2000, 38, 362–368. [Google Scholar] [CrossRef]
- Puri, A.; Bajpai, S.; Meredith, S.; Aravind, L.; Krause, P.J.; Kumar, S. Babesia microti: Pathogen genomics, genetic variability, immunodominant antigens, and pathogenesis. Front. Microbiol. 2021, 12, 697669. [Google Scholar] [CrossRef]
- World Malaria Report 2020: 20 Years of Global Progress and Challenges; World Health Organization: Geneva, Switzerland, 2020.
- Berhane, A.; Anderson, K.; Mihreteab, S.; Gresty, K.; Rogier, E.; Mohamed, S.; Hagos, F.; Embaye, G.; Chinorumba, A.; Zehaie, A.; et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, eritrea. Emerg. Infect. Dis. 2018, 24, 462–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pati, P.; Dhangadamajhi, G.; Bal, M.; Ranjit, M. High proportions of pfhrp2 gene deletion and performance of HRP2-based rapid diagnostic test in Plasmodium falciparum field isolates of Odisha. Malar. J. 2018, 17, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leeflang, P.; Perié, N.M. Comparative immunofluorescent studies on 4 Babesia species of cattle. Res. Vet. Sci. 1972, 13, 342–346. [Google Scholar] [CrossRef]
- Goldman, M.; Pipano, E.; Rosenberg, A.S. Fluorescent antibody tests for Babesia bigemina and B. berbera. Res. Vet. Sci. 1972, 13, 77–81. [Google Scholar] [CrossRef]
- Cox, F.E.; Turner, S.A. Antigenic relationships between the malaria parasites and piroplasms of mice as determined by the fluorescent-antibody technique. Bull. World Health Organ. 1970, 43, 337–340. [Google Scholar]
- Chisholm, E.S.; RuebushIi, T.K., 2nd; Sulzer, A.J.; Healy, G.R. Babesia microti infection in man: Evaluation of an indirect immunofluorescent antibody test. Am. J. Trop. Med. Hyg. 1978, 27, 14–19. [Google Scholar] [CrossRef]
- Boustani, M.R.; Gelfand, J.A. Babesiosis. Clin. Infect. Dis. 1996, 22, 611–615. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention. Babesiosis (Babesia spp.) 2011 CASE definition. Available online: https://ndc.services.cdc.gov/case-definitions/babesiosis-2011/ (accessed on 30 September 2021).
- Tonnetti, L.; Johnson, S.; Cable, R.; Rios, J.; Spencer, B.; Leiby, D. Natural history study (NHS) of Babesia microti in Connecticut blood donors. Transfusion 2009, 49, 35A–36A. [Google Scholar]
- Ruebush, T.K., 2nd; Chisholm, E.S.; Sulzer, A.J.; Healy, G.R.; Ii, T.K.R. Development and persistence of antibody in persons infected with Babesia microti. Am. J. Trop. Med. Hyg. 1981, 30, 291–292. [Google Scholar] [CrossRef]
- Loa, C.C.; Adelson, M.E.; Mordechai, E.; Raphaelli, I.; Tilton, R.C. Serological diagnosis of human babesiosis by IgG enzyme-linked immunosorbent assay. Curr. Microbiol. 2004, 49, 385–389. [Google Scholar] [CrossRef]
- Meeusen, E.; Lloyd, S.; Soulsby, E. Antibody levels in adoptively immunized mice after infection with Babesia microti or injection with antigen fractions. Aust. J. Exp. Biol. Med. Sci. 1985, 63 Pt 3, 261–272. [Google Scholar] [CrossRef]
- Houghton, R.L.; Homer, M.J.; Reynolds, L.D.; Sleath, P.R.; Lodes, M.J.; Berardi, V.; Leiby, D.A.; Persing, D.H. Identification of Babesia microti-specific immunodominant epitopes and development of a peptide EIA for detection of antibodies in serum. Transfusion 2002, 42, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.; Chieves, L.; Johnson, G.; Buening, G. Multiplex polymerase chain reaction based assay for the detection of Babesia bigemina, Babesia bovis and Anaplasma marginale DNA in bovine blood. Vet. Parasitol. 1993, 50, 69–81. [Google Scholar] [CrossRef]
- Parodi, P.; Corbellini, L.G.; Leotti, V.B.; Rivero, R.; Miraballes, C.; Riet-Correa, F.; Venzal, J.M.; Armúa-Fernández, M.T. Validation of a multiplex PCR assay to detect Babesia spp. and Anaplasma marginale in cattle in Uruguay in the absence of a gold standard test. J. Vet. Diagn. Investig. 2020, 33, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, I.; Burri, C.; Noda, A.; Douet, V.; Gern, L. Multiplex PCR for molecular screening of Borrelia burgdorferi sensu lato, Anaplasma spp. and Babesia spp. Ann. Agric. Environ. Med. 2015, 22, 642–646. [Google Scholar] [CrossRef] [Green Version]
- Tokarz, R.; Tagliafierro, T.; Cucura, D.M.; Rochlin, I.; Sameroff, S.; Lipkin, W.I. Detection of Anaplasma phagocytophilum, Babesia microti, Borrelia burgdorferi, Borrelia miyamotoi, and Powassan Virus in ticks by a multiplex real-time reverse transcription-PCR assay. mSphere 2017, 2, e00151-17. [Google Scholar] [CrossRef] [Green Version]
- Buchan, B.W.; Jobe, D.A.; Mashock, M.; Gerstbrein, D.; Faron, M.L.; Ledeboer, N.A.; Callister, S.M. Evaluation of a novel multiplex high-definition PCR assay for detection of tick-borne pathogens in whole-blood specimens. J. Clin. Microbiol. 2019, 57, e00513-19. [Google Scholar] [CrossRef] [Green Version]
- Livengood, J.; Hutchinson, M.L.; Thirumalapura, N.; Tewari, D. Detection of Babesia, Borrelia, Anaplasma, and Rickettsia spp. in adult black-legged ticks (Ixodes scapularis) from Pennsylvania, United States, with a luminex multiplex bead assay. Vector Borne Zoonotic Dis. 2020, 20, 406–411. [Google Scholar] [CrossRef]
- Priest, J.W.; Moss, D.M.; Won, K.; Todd, C.W.; Henderson, L.; Jones, C.C.; Wilson, M. Multiplex assay detection of immunoglobulin G antibodies that recognize Babesia microti antigens. Clin. Vaccine Immunol. 2012, 19, 1539–1548. [Google Scholar] [CrossRef] [Green Version]
- Duh, D.; Jelovšek, M.; Županc, T.A. Evaluation of an indirect fluorescence immunoassay for the detection of serum antibodies against Babesia divergens in humans. Parasitology 2006, 134, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Persing, D.H.; Herwaldt, B.L.; Glaser, C.; Lane, R.S.; Thomford, J.W.; Mathiesen, D.; Krause, P.J.; Phillip, D.F.; Conrad, P.A. Infection with a babesia-like organism in Northern California. N. Engl. J. Med. 1995, 332, 298–303. [Google Scholar] [CrossRef]
- Herwaldt, B.L.; Persing, D.H.; Precigout, E.A.; Goff, W.L.; Mathiesen, D.A.; Taylor, P.W.; Eberhard, M.L.; Gorenflot, A.F. A fatal case of babesiosis in Missouri: Identification of another piroplasm that infects humans. Ann. Intern. Med. 1996, 124, 643–650. [Google Scholar] [CrossRef]
- Herwaldt, B.L.; de Bruyn, G.; Pieniazek, N.J.; Homer, M.; Lofy, K.H.; Slemenda, S.B.; Fritsche, T.R.; Persing, D.H.; Limaye, A.P. Babesia divergens–like Infection, Washington State. Emerg. Infect. Dis. 2004, 10, 622–629. [Google Scholar] [CrossRef]
- Wilson, M.; Naccache, S.N.; Samayoa, E.; Biagtan, M.; Bashir, H.; Yu, G.; Salamat, S.M.; Somasekar, S.; Federman, S.; Miller, S.; et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 2014, 370, 2408–2417. [Google Scholar] [CrossRef] [Green Version]
- Doan, T.; Wilson, M.R.; Crawford, E.D.; Chow, E.D.; Khan, L.M.; Knopp, K.A.; O’Donovan, B.D.; Xia, D.; Hacker, J.K.; Stewart, J.M.; et al. Illuminating uveitis: Metagenomic deep sequencing identifies common and rare pathogens. Genome Med. 2016, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Greninger, A.L.; Naccache, S.N.; Federman, S.; Yu, G.; Mbala, P.; Bres, V.; Stryke, D.; Bouquet, J.; Somasekar, S.; Linnen, J.M.; et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, C.H.; Hennelly, C.M.; Lin, J.T.; Ubalee, R.; Boyce, R.M.; Mulogo, E.M.; Hathaway, N.; Thwai, K.L.; Phanzu, F.; Kalonji, A.; et al. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. EBioMedicine 2021, 68, 103415. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Hunt, J.V.; Kosinski, K.S.; Demirjian, Z.N.; Huggins, C.; Etkind, P.; Marcus, L.C.; Spielman, A. Treatment of transfusion-transmitted babesiosis by exchange transfusion. N. Engl. J. Med. 1980, 303, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Herwaldt, B.L.; Linden, J.V.; Bosserman, E.; Young, C.; Olkowska, D.; Wilson, M. Transfusion-associated babesiosis in the United States: A description of cases. Ann. Intern. Med. 2011, 155, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linden, J.V.; Prusinski, M.; Crowder, L.; Tonnetti, L.; Stramer, S.L.; Kessler, D.A.; White, J.; Shaz, B.; Olkowska, D. Transfusion-transmitted and community-acquired babesiosis in New York, 2004 to 2015. Transfusion 2018, 58, 660–668. [Google Scholar] [CrossRef]
- Mintz, E.D.; Anderson, J.F.; Cable, R.G.; Hadler, J.L. Transfusion-transmitted babesiosis: A case report from a new endemic area. Transfusion 1991, 31, 365–368. [Google Scholar] [CrossRef]
- Menis, M.; Forshee, R.A.; Kumar, S.; Mckean, S.; Warnock, R.; Izurieta, H.S.; Gondalia, R.; Johnson, C.; Mintz, P.D.; Walderhaug, M.O.; et al. Babesiosis occurrence among the elderly in the United States, as recorded in large medicare databases during 2006–2013. PLoS ONE 2015, 10, e0140332. [Google Scholar] [CrossRef]
- Johnson, S.T.; Cable, R.G.; Tonnetti, L.; Spencer, B.; Rios, J.; Leiby, D.A. Transfusion complications: Seroprevalence of Babesia microti in blood donors from Babesia-endemic areas of the northeastern United States: 2000 through 2007. Transfusion 2009, 49, 2574–2582. [Google Scholar] [CrossRef]
- Leiby, D.A.; Chung, A.P.; Gill, J.E.; Houghton, R.L.; Persing, D.H.; Badon, S.; Cable, R.G. Demonstrable parasitemia among Connecticut blood donors with antibodies to Babesia microti. Transfusion 2005, 45, 1804–1810. [Google Scholar] [CrossRef] [PubMed]
- Leiby, D.A.; Johnson, S.T.; Won, K.Y.; Nace, E.K.; Slemenda, S.B.; Pieniazek, N.J.; Cable, R.G.; Herwaldt, B.L. A longitudinal study of Babesia microti infection in seropositive blood donors. Transfusion 2014, 54, 2217–2225. [Google Scholar] [CrossRef] [Green Version]
- Levin, A.E.; Williamson, P.C.; Bloch, E.M.; Clifford, J.; Cyrus, S.; Shaz, B.; Kessler, D.; Gorlin, J.; Erwin, J.L.; Krueger, N.X.; et al. Serologic screening of United States blood donors for Babesia microti using an investigational enzyme immunoassay. Transfusion 2016, 56, 1866–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cangelosi, J.J.; Sarvat, B.; Sarria, J.C.; Herwaldt, B.L.; Indrikovs, A.J. Transmission of Babesia microti by blood transfusion in Texas. Vox Sang. 2008, 95, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.; Civen, R. Babesiosis acquired through blood transfusion, California, USA. Emerg. Infect. Dis. 2009, 15, 785–787. [Google Scholar] [CrossRef] [PubMed]
- Expert Committee on Biological Standardization. A collaborative study to evaluate the proposed 6th WHO International Standard for Hepatitis C Virus (HCV) RNA for nucleic acid amplification techniques (NAT). World Health Organization. 2019. Available online: https://www.who.int/biologicals/expert_committee/BS.2019.2358_6thHCVIS_WHOECBS_July2019.pdf (accessed on 21 November 2021).
- Expert Committee on Biological Standardization. International collaborative study to establish the 4th WHO International Standard for HIV-1 NAT assays. World Health Organization. 2017. Available online: https://apps.who.int/iris/handle/10665/260256 (accessed on 21 November 2021).



| Method | Target | Limit of Detection | Reference |
|---|---|---|---|
| Blood Film | 20–100 pRBCs */μL | [28,29,30] | |
| Experimental Inoculation | 63 pRBCs/inoculation [into mice] | [46] | |
| Fluorescent Nucleic Acid Probes | 100 pg DNA (~30 parasites)[B. bovis **] | [47] | |
| PCR | 18S rRNA | 3 parasites/50 μL | [33] |
| RT-PCR | 18S rRNA | 12.92 parasites/2 mL | [40] |
| BMN genes | 10 pRBCs/mL | [45] | |
| ddPCR | 18S rRNA | 10 copies | [41] |
| TMA | 18S rRNA | 3 pRBCs/mL | [35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meredith, S.; Oakley, M.; Kumar, S. Technologies for Detection of Babesia microti: Advances and Challenges. Pathogens 2021, 10, 1563. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens10121563
Meredith S, Oakley M, Kumar S. Technologies for Detection of Babesia microti: Advances and Challenges. Pathogens. 2021; 10(12):1563. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens10121563
Chicago/Turabian StyleMeredith, Scott, Miranda Oakley, and Sanjai Kumar. 2021. "Technologies for Detection of Babesia microti: Advances and Challenges" Pathogens 10, no. 12: 1563. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens10121563