A Global Analysis of Enzyme Compartmentalization to Glycosomes
Abstract
:1. Introduction
2. Results
2.1. Meta-Analysis of Existing Studies
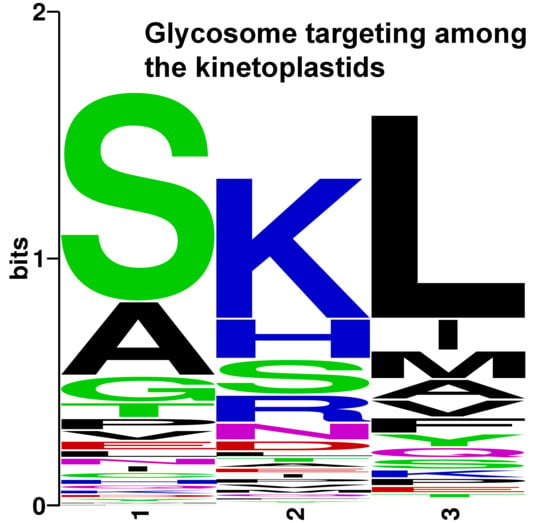
2.2. An Adequate Algorithm for Glycosome Localization
2.3. Glycosome Localization Prediction of Orthologues of Gcec Proteins across Kinetoplastids
2.4. Utilizing the PTS1 Signal Algorithm in Other Datasets
2.5. Similarities of Protein Compositions of the Glycosome with the Peroxisomes and the Mitochondrion
3. Discussion
4. Materials and Methods
4.1. Scoring of Meta-Analysis
4.2. Categorization of Proteins into Metabolic Pathways
4.3. Identification of Orthologues in Other Organisms
4.4. Development of Glycosome Targeting Signal 1 (PTS1) Algorithm
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ferguson, M.A.; Low, M.G.; Cross, G.A. Glycosyl-sn-1,2-dimyristylphosphatidylinositol is covalently linked to Trypanosoma brucei variant surface glycoprotein. J. Biol. Chem. 1985, 260, 14547–14555. [Google Scholar] [PubMed]
- Imboden, M.A.; Laird, P.W.; Affolter, M.; Seebeck, T. Transcription of the intergenic regions of the tubulin gene cluster of Trypanosoma brucei: Evidence for a polycistronic transcription unit in a eukaryote. Nucleic Acids Res. 1987, 15, 7357–7368. [Google Scholar] [CrossRef] [PubMed]
- Muhich, M.L.; Boothroyd, J.C. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: Evidence for a precursor role in mRNA synthesis. Mol. Cell. Biol. 1988, 8, 3837–3846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, R.E.; Boothroyd, J.C. Evidence for Trans splicing in trypanosomes. Cell 1986, 47, 527–535. [Google Scholar] [CrossRef]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Lukeš, J.; Skalický, T.; Týč, J.; Votýpka, J.; Yurchenko, V. Evolution of parasitism in kinetoplastid flagellates. Mol. Biochem. Parasitol. 2014, 195, 115–122. [Google Scholar] [CrossRef]
- Allmann, S.; Bringaud, F. Glycosomes: A comprehensive view of their metabolic roles in T. brucei. Int. J. Biochem. Cell Biol. 2017, 85, 85–90. [Google Scholar] [CrossRef]
- Moyersoen, J.; Choe, J.; Fan, E.; Hol, W.G.J.; Michels, P.A.M. Biogenesis of peroxisomes and glycosomes: Trypanosomatid glycosome assembly is a promising new drug target. FEMS Microbiol. Rev. 2004, 28, 603–643. [Google Scholar] [CrossRef] [Green Version]
- Szöör, B.; Haanstra, J.R.; Gualdrón-López, M.; Michels, P.A. Evolution, dynamics and specialized functions of glycosomes in metabolism and development of trypanosomatids. Curr. Opin. Microbiol. 2014, 22, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Jamdhade, M.D.; Pawar, H.; Chavan, S.; Sathe, G.; Umasankar, P.K.; Mahale, K.N.; Dixit, T.; Madugundu, A.K.; Prasad, T.S.K.; Gowda, H.; et al. Comprehensive proteomics analysis of glycosomes from Leishmania donovani. OMICS 2015, 19, 157–170. [Google Scholar] [CrossRef] [Green Version]
- Acosta, H.; Burchmore, R.; Naula, C.; Gualdrón-López, M.; Quintero-Troconis, E.; Cáceres, A.J.; Michels, P.A.M.; Concepción, J.L.; Quiñones, W. Proteomic analysis of glycosomes from Trypanosoma cruzi epimastigotes. Mol. Biochem. Parasitol. 2019, 229, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Güther, M.L.S.; Urbaniak, M.D.; Tavendale, A.; Prescott, A.; Ferguson, M.A.J. High-Confidence Glycosome Proteome for Procyclic Form Trypanosoma brucei by Epitope-Tag Organelle Enrichment and SILAC Proteomics. J. Proteome Res. 2014, 13, 2796–2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opperdoes, F.R.; Szikora, J.P. In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol. Biochem. Parasitol. 2006, 147, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Makiuchi, T.; Annoura, T.; Hashimoto, M.; Hashimoto, T.; Aoki, T.; Nara, T. Compartmentalization of a Glycolytic Enzyme in Diplonema, a Non-kinetoplastid Euglenozoan. Protist 2011, 162, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Hashimoto, M.; Williams, T.A.; Hirawake-Mogi, H.; Makiuchi, T.; Tsubouchi, A.; Kaga, N.; Taka, H.; Fujimura, T.; Koike, M.; et al. Differential remodelling of peroxisome function underpins the environmental and metabolic adaptability of diplonemids and kinetoplastids. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, S.R.; Prathalingam, S.R.; Taylor, M.C.; Ahmed, A.; Horn, D.; Kelly, J.M. Functional characterisation of the iron superoxide dismutase gene repertoire in Trypanosoma brucei. Free Radic. Biol. Med. 2006, 40, 198–209. [Google Scholar] [CrossRef]
- Dufernez, F.; Yernaux, C.; Gerbod, D.; Noël, C.; Chauvenet, M.; Wintjens, R.; Edgcomb, V.P.; Capron, M.; Opperdoes, F.R.; Viscogliosi, E. The presence of four iron-containing superoxide dismutase isozymes in Trypanosomatidae: Characterization, subcellular localization, and phylogenetic origin in Trypanosoma brucei. Free Radic. Biol. Med. 2006, 40, 210–225. [Google Scholar] [CrossRef]
- Sargsyan, Y.; Thoms, S. Staying in Healthy Contact: How Peroxisomes Interact with Other Cell Organelles. Trends Mol. Med. 2020, 26, 201–214. [Google Scholar] [CrossRef]
- Schlüter, A.; Real-Chicharro, A.; Gabaldón, T.; Sánchez-Jiménez, F.; Pujol, A. PeroxisomeDB 2.0: An integrative view of the global peroxisomal metabolome. Nucleic Acids Res. 2010, 38, D800-5. [Google Scholar]
- Colasante, C.; Ellis, M.; Ruppert, T.; Voncken, F. Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 2006, 6, 3275–3293. [Google Scholar] [CrossRef]
- Parsons, M.; Nielsen, B. Trypanosoma brucei: Two-dimensional gel analysis of the major glycosomal proteins during the life cycle. Exp. Parasitol. 1990, 70, 276–285. [Google Scholar] [CrossRef]
- Dean, S.; Sunter, J.D.; Wheeler, R.J. TrypTag.org: A Trypanosome Genome-wide Protein Localisation Resource. Trends Parasitol. 2017, 33, 80–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, J.C.; Bomgarden, R.D. Sample preparation for mass spectrometry-based proteomics; from proteomes to peptides. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2016; Volume 919, pp. 43–62. [Google Scholar]
- Halliday, C.; Billington, K.; Wang, Z.; Madden, R.; Dean, S.; Sunter, J.D.; Wheeler, R.J. Cellular landmarks of Trypanosoma brucei and Leishmania mexicana. Mol. Biochem. Parasitol. 2019, 230, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, D.J.; Gutteridge, W.E.; Opperdoes, F.R. A novel location for two enzymes of de novo pyrimidine biosynthesis in trypanosomes and Leishmania. FEBS Lett. 1981, 128, 27–29. [Google Scholar] [CrossRef] [Green Version]
- Szöor, B.; Ruberto, I.; Burchmore, R.; Matthews, K.R. A novel phosphatase cascade regulates differentiation in Trypanosoma brucei via a glycosomal signaling pathway. Genes Dev. 2010, 24, 1306–1316. [Google Scholar] [CrossRef] [Green Version]
- Aranovich, A.; Hua, R.; Rutenberg, A.D.; Kim, P.K. PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 2014, 127, 3675–3686. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, Y.; Gao, C.; Jiang, L.; Guo, D. PPero, a Computational Model for Plant PTS1 Type Peroxisomal Protein Prediction. PLoS ONE 2017, 12, e0168912. [Google Scholar] [CrossRef] [PubMed]
- Neuberger, G.; Maurer-Stroh, S.; Eisenhaber, B.; Hartig, A.; Eisenhaber, F. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 2003, 328, 581–592. [Google Scholar] [CrossRef]
- Fox, T.D. Mitochondrial protein synthesis, import, and assembly. Genetics 2012, 192, 1203–1234. [Google Scholar] [CrossRef] [Green Version]
- Kalel, V.C.; Mäser, P.; Sattler, M.; Erdmann, R.; Popowicz, G.M. Come, sweet death: Targeting glycosomal protein import for antitrypanosomal drug development. Curr. Opin. Microbiol. 2018, 46, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, A.; Mattie, S.; Prudent, J.; Mcbride, H.M. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 2017, 542, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.T.; McQueeney, K.E.; Patel, T.; Morris, M.T. Localization of a Trypanosome Peroxin to the Endoplasmic Reticulum. J. Eukaryot. Microbiol. 2017, 64, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, R.; Reumann, S.; Lisik, P.; Tietz, S.; Olsen, L.J.; Hu, J. Proteome analysis of peroxisomes from dark-treated senescent Arabidopsis leaves. J. Integr. Plant Biol. 2018, 60, 1028–1050. [Google Scholar] [CrossRef] [Green Version]
- Gronemeyer, T.; Wiese, S.; Ofman, R.; Bunse, C.; Pawlas, M.; Hayen, H.; Eisenacher, M.; Stephan, C.; Meyer, H.E.; Waterham, H.R.; et al. The Proteome of Human Liver Peroxisomes: Identification of Five New Peroxisomal Constituents by a Label-Free Quantitative Proteomics Survey. PLoS ONE 2013, 8, e57395. [Google Scholar] [CrossRef]
- Kerkhoven, E.J.; Achcar, F.; Alibu, V.P.; Burchmore, R.J.; Gilbert, I.H.; Trybiło, M.; Driessen, N.N.; Gilbert, D.; Breitling, R.; Bakker, B.M.; et al. Handling Uncertainty in Dynamic Models: The Pentose Phosphate Pathway in Trypanosoma brucei. PLoS Comput. Biol. 2013, 9, e1003371. [Google Scholar] [CrossRef] [Green Version]
- Becco, L.; Smircich, P.; Garat, B. Conserved motifs in nuclear genes encoding predicted mitochondrial proteins in Trypanosoma cruzi. PLoS ONE 2019, 14, e0215160. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Cui, J.; Nilsson, D.; Gunasekera, K.; Chanfon, A.; Song, X.; Wang, H.; Xu, Y.; Ochsenreiter, T. The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res. 2010, 38, 7378–7387. [Google Scholar] [CrossRef] [Green Version]
- Horáková, E.; Faktorová, D.; Kraeva, N.; Kaur, B.; Van Den Abbeele, J.; Yurchenko, V.; Lukeš, J. Catalase compromises the development of the insect and mammalian stages of Trypanosoma brucei. FEBS J. 2019, 287, 964–977. [Google Scholar] [CrossRef]
- Bauer, S.; Morris, J.C.; Morris, M.T. Environmentally regulated glycosome protein composition in the African trypanosome. Eukaryot. Cell 2013, 12, 1072–1079. [Google Scholar] [CrossRef] [Green Version]
- Herman, M.; Pérez-Morga, D.; Schtickzelle, N.; Michels, P.A.M. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy 2008, 4, 294–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tielens, A.G.; van Hellemond, J.J. Surprising variety in energy metabolism within Trypanosomatidae. Trends Parasitol. 2009, 25, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Trindade, S.; Rijo-Ferreira, F.; Carvalho, T.; Pinto-Neves, D.; Guegan, F.; Aresta-Branco, F.; Bento, F.; Young, S.A.; Pinto, A.; Van Den Abbeele, J.; et al. Trypanosoma brucei Parasites Occupy and Functionally Adapt to the Adipose Tissue in Mice. Cell Host Microbe 2016, 19, 837–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flegontov, P.; Butenko, A.; Firsov, S.; Kraeva, N.; Eliáš, M.; Field, M.C.; Filatov, D.; Flegontova, O.; Gerasimov, E.S.; Hlaváčová, J.; et al. Genome of Leptomonas pyrrhocoris: A high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci. Rep. 2016, 6, 23704. [Google Scholar] [CrossRef]
- Freitag, J.; Ast, J.; Bölker, M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 2012, 485, 522–525. [Google Scholar] [CrossRef]
- Ebenezer, T.G.E.; Carrington, M.; Lebert, M.; Kelly, S.; Field, M.C. Euglena gracilis genome and transcriptome: Organelles, nuclear genome assembly strategies and initial features. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2017; Volume 979, pp. 125–140. [Google Scholar]
- Ebenezer, T.E.; Zoltner, M.; Burrell, A.; Nenarokova, A.; Novák Vanclová, A.M.G.; Prasad, B.; Soukal, P.; Santana-Molina, C.; O’Neill, E.; Nankissoor, N.N.; et al. Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biol. 2019, 17, 11. [Google Scholar] [CrossRef] [Green Version]
- Kaur, B.; Valach, M.; Peña-Diaz, P.; Moreira, S.; Keeling, P.J.; Burger, G.; Lukeš, J.; Faktorová, D. Transformation of Diplonema papillatum, the type species of the highly diverse and abundant marine microeukaryotes Diplonemida (Euglenozoa). Environ. Microbiol. 2018, 20, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, Y.; Tomiyama, T.; Maruta, T.; Tomita, M.; Ishikawa, T.; Arakawa, K. De novo assembly and comparative transcriptome analysis of Euglena gracilis in response to anaerobic conditions. BMC Genom. 2016, 17, 182. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]





| KEGG Reference Ortholog | Major KEGG Pathway | Name of Protein | PTS1/PTS2 |
|---|---|---|---|
| K18561 | Glycolysis | NADH-dependent fumarate reductase | PTS1 |
| Glycolysis | UDP-glc 4’-epimerase | PTS1 | |
| K00850 | Glycolysis | ATP-dependent 6-phosphofructokinase, glycosomal | PTS1 |
| K00844 | Glycolysis/gluconeogenesis | Hexokinase 1 | PTS2 |
| K01810 | Glycolysis/gluconeogenesis | Glucose-6-phosphate isomerase | PTS1 |
| K00134 | Glycolysis/gluconeogenesis | Glyceraldehyde 3-phosphate dehydrogenase | PTS1 |
| K01006 | Glycolysis/gluconeogenesis | Pyruvate phosphate dikinase | PTS1 |
| K00927 | Glycolysis/gluconeogenesis | Phosphoglycerate kinase | PTS2 |
| K01792 | Glycolysis/gluconeogenesis | Aldose 1-epimerase | PTS1 |
| K01623 | Glycolysis/gluconeogenesis | Fructose-bisphosphate aldolase | |
| K01803 | Glycolysis/gluconeogenesis | Triose phosphate isomerase | |
| K03841 | Gluconeogenesis | Fructose-1,6-bisphosphatase | PTS1 |
| K01610 | Gluconeogenesis | Phosphoenolpyruvate carboxykinase [ATP] | PTS1 |
| K00026 | Gluconeogenesis | Glycosomal malate dehydrogenase | PTS1 |
| Gluconeogenesis | UDP-glucose pyrophosphorylase | ||
| K00760 | Purine metabolism | Hypoxanthine-guanine phosphoribosyltransferase 1 | PTS1 |
| K00760 | Purine metabolism | Hypoxanthine-guanine phosphoribosyltransferase 2 | PTS1 |
| K00088 | Purine metabolism | Inosine-5’-monophosphate dehydrogenase | PTS1 |
| K00942 | Purine metabolism | Guanylate kinase | PTS1 |
| K00759 | Purine metabolism | Adenine phosphoribosyltransferase | PTS1 |
| K01490 | Purine metabolism | AMP deaminase | PTS1 |
| K00939 | Purine metabolism | Adenylate kinase | PTS1 |
| K00088 | Purine metabolism | Guanosine monophosphate reductase | PTS1 |
| K00036 | Pentose phosphate pathway | Glucose-6-phosphate 1-dehydrogenase (G6PD) | |
| K00852 | Pentose phosphate pathway | Ribokinase | |
| K01100 | Pentose phosphate pathway | Sedoheptulose-1,7-bisphosphatase | PTS1 |
| K01619 | Pentose phosphate pathway | Deoxyribose-phosphate aldolase | PTS1 |
| K00615 | Pentose phosphate pathway | Transketolase | PTS1 |
| K01057 | Pentose phosphate pathway | 6-phosphogluconolactonase | |
| K00864 | Glycerophospholipid metabolism | Glycerol kinase | PTS1 |
| K00803 | Glycerophospholipid metabolism | Alkyl-dihydroxyacetone phosphate synthase | PTS1 |
| K00649 | Glycerophospholipid metabolism | Dihydroxyacetonephosphate acyltransferase | PTS1 |
| K00006 | Glycerophospholipid metabolism | Glycerol-3-phosphate dehydrogenase (NAD(+)) | PTS1 |
| K00022 | Fatty acid metabolism | Enoyl-CoA hydratase/Enoyl-CoA isomerase/3- hydroxyacyl-CoA dehydrogenase | PTS2 |
| K08766 | Fatty acid metabolism | Carnitine O-palmitoyltransferase | PTS1 |
| K11262 | Fatty acid metabolism | Acetyl-CoA carboxylase | |
| K11207 | Redox maintenance | Trypanothione/tryparedoxin dependent peroxidase 2 | |
| K01833 | Redox maintenance | Trypanothione synthetase | |
| K00103 | Redox maintenance | L-galactonolactone oxidase | PTS1 |
| K00869 | Terpenoid biosynthesis | Mevalonate kinase | PTS1 |
| K01823 | Terpenoid biosynthesis | Isopentenyl-diphosphate delta-isomerase (type II) (idi1) | |
| K00031 | TCA cycle/glutathione metabolism | Isocitrate dehydrogenase | PTS1 |
| K01438 | Amino acid biosynthesis (arginine) | Acetylornithine deacetylase | PTS1 |
| K01107 | Insositol phosphate metabolism | Inositol polyphosphate 1-phosphatase | |
| K13421 | Pyrimidine metabolism | Orotidine-5-phosphate decarboxylase/Orotate phosphoribosyltransferase | PTS1 |
| K15731 | RNA polymerase II C-terminal domain phosphatase | PTP1-interacting protein, 39 kDa/TFIIF-stimulated CTD phosphatase | PTS1 |
| K09829 | Steroid biosynthesis (ERG2) | C-8 sterol isomerase-like protein | PTS1 |
| K10703 | Long chain fatty acid synthesis | Protein tyrosine phosphatase | |
| N/A | N/A | Thymine-7-hydroxylase | PTS1 |
| N/A | N/A | Hypothetical protein (Q580K0) | PTS1 |
| N/A | N/A | Hypothetical protein (Q389Y7) | |
| N/A | N/A | Hypothetical protein (Q38C56) | |
| N/A | N/A | Hypothetical protein (Q386P8) | PTS1 |
| N/A | N/A | Hypothetical protein(Q388J7) | |
| N/A | N/A | Hypothetical protein (Q38DM9) | |
| N/A | N/A | Hypothetical protein (Q383Q3) | |
| N/A | N/A | Hypothetical protein (Q38AC3) |
| KEGG Reference Ortholog | Major KEGG Pathway | Name of Protein | PTS1/PTS2 |
|---|---|---|---|
| K00845 | Glycolysis | Glucokinase | PTS1 |
| K01809 | Glycolysis/gluconeogenesis | Phosphomannose isomerase | PTS1 |
| K17497 | Glycolysis/gluconeogenesis | Phosphomannomutase-like protein | PTS1 |
| K00849 | Glycolysis/gluconeogenesis | Galactokinase-like protein | PTS1 |
| K02564 | Glycolysis/gluconeogenesis | Glucosamine-6-phosphate isomerase | PTS1 |
| K00927 | Glycolysis/gluconeogenesis | Pas-domain containing phosphoglycerate kinase | PTS1 |
| K01443 | Glycolysis/gluconeogenesis | N-acetylglucosamine-6-phosphate deacetylase-like protein | PTS1 |
| K00863 | Glycolysis/gluconeogenesis | Dihydroxyacetone kinase 1-like | PTS1 |
| K01518 | Purine metabolism | Kinetoplastid-specific phospho-protein phosphatase | PTS1 |
| K00759 | Purine metabolism | Adenine phosphoribosyltransferase | PTS1 |
| K00853 | Pentose phosphate pathway | L-ribulokinase | PTS1 |
| K06128 | Glycerophospholipid metabolism | Lysophospholipase | PTS1 |
| Fatty acid metabolism | Acyl-CoA binding protein | PTS1 | |
| K00311 | Fatty acid metabolism | Electron transfer flavoprotein-ubiquinone oxidoreductase | PTS1 |
| K13356 | Fatty acid metabolism | Fatty acyl- CoA reducatase | PTS1 |
| K00632 | Fatty acid metabolism | 3-ketoacyl- CoA thiolase | PTS2 |
| Fatty acid epoxide hydrolase | Epoxide hydrolase | PTS1 | |
| K04283 | Redox maintenance | Trypanothione-disulfide reductase | PTS1 |
| K11185 | Redox maintenance | Tryparedoxin peroxidase | PTS1 |
| Redox maintenance | Dj-1 family protein | PTS1 | |
| K04564 | Redox maintenance | Iron superoxide dismutase | PTS1 |
| K04564 | Redox maintenance | Iron superoxide dismutase | PTS1 |
| Redox maintenance | 2-oxoglutarate (2og) and Fe(II)-dependent oxygenase superfamily protein | PTS1 | |
| K01940 | Urea cycle | Arginino-succinate synthase | PTS1 |
| K01438 | Urea cycle | Acetylornithine deacetylase-like | PTS1 |
| K01745 | Amino acid degradation | Histidine ammonia-lyase | PTS1 |
| K02614 | Amino acid degradation | Thioesterase-like superfamily | PTS1 |
| Peptide cleavage | Peptidase T | PTS2 | |
| Protein cleavage | Carboxypeptidase M32 | PTS2 | |
| K02150 | pH regulation | V-ATPase, subunit E | PTS1 |
| Pyrophosphate and poly phosphate metabolism | Acidocalcisomal exopolyphosphatase | PTS1 | |
| K02218 | Signal pathway regulation | Casein kinase I, isoform 2 | PTS2 |
| K01676 | TCA cycle | Fumarate hydratase, class I (FHM) | PTS2 |
| K00972 | Amino and nucleotide sugar metabolism | UDP-N-acetylglucosamine pyrophosphorylase | PTS1 |
| N/A | Hypothetical protein (Q4DBW4) | PTS1 | |
| N/A | Hypothetical protein (Q57TT5) | PTS1 | |
| N/A | Hypothetical protein (Q381V8) | PTS1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durrani, H.; Hampton, M.; Rumbley, J.N.; Zimmer, S.L. A Global Analysis of Enzyme Compartmentalization to Glycosomes. Pathogens 2020, 9, 281. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9040281
Durrani H, Hampton M, Rumbley JN, Zimmer SL. A Global Analysis of Enzyme Compartmentalization to Glycosomes. Pathogens. 2020; 9(4):281. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9040281
Chicago/Turabian StyleDurrani, Hina, Marshall Hampton, Jon N. Rumbley, and Sara L. Zimmer. 2020. "A Global Analysis of Enzyme Compartmentalization to Glycosomes" Pathogens 9, no. 4: 281. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9040281