Biofilm Formation Ability and Presence of Adhesion Genes among Coagulase-Negative and Coagulase-Positive Staphylococci Isolates from Raw Cow’s Milk
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Isolation of Staphylococci Strains
4.2. Detection of the Ability to Slime Production by Congo Red Agar (CRA) Method
4.3. Biofilm Forming Ability Detection by Microtiter Plate Method (MTP)
4.4. Detection of Biofilm-Associated Genes
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cunha, R.C.; Rosa, M.D.H.D.; Silva, C.D.; Santos, F.D.S.; Leite, F.P.L. Staphylococcal slime layers and biofilm from different origins. Ciência Rural 2019, 49, e20180783. [Google Scholar] [CrossRef]
- Coorevits, A.; De Jonghe, V.; Vandroemme, J.; Reekmans, R.; Heyrman, J.; Messens, W.; De Vos, P.; Heyndrickx, M. Comparative analysis of the diversity of aerobic spore-forming bacteria in raw milk from organic and conventional dairy farms. Syst. Appl. Microbiol. 2008, 31, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Vacheyrou, M.; Normand, A.C.; Guyot, P.; Cassagne, C.; Piarroux, R.; Bouton, Y. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int. J. Food Microbiol. 2011, 146, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Baranowska, A.; Chojnowski, W.; Nowak, H. Dezynfekcja w zakładach mleczarskich. Eng. Sci. Technol. 2014, 4, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Daniel, R.M.; Coolbear, T. Detection and impact of protease and lipase activities in milk and milk powders. Int. Dairy J. 2003, 13, 255–275. [Google Scholar] [CrossRef]
- Verdier-Metz, I.; Gagne, G.; Bornes, S.; Monsallier, F.; Veisseire, P.; Delbès-Paus, C.; Montel, M.C. Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl. Environ. Microbiol. 2012, 78, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Zhu, X. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 2009, 20, 407–413. [Google Scholar] [CrossRef]
- Turchi, B.; Bertelloni, F.; Marzoli, F.; Cerri, D.; Tola, S.; Azara, E.; Longheu, C.M.; Tassi, R.; Schiavo, M.; Cilia, G.; et al. Coagulase negative staphylococci from ovine milk: Genotypic and phenotypic characterization of susceptibility to antibiotics, disinfectants and biofilm production. Small Rumin. Res. 2020, 183, 106030. [Google Scholar] [CrossRef]
- Kukhtyn, M.; Berhilevych, O.; Kravcheniuk, K.; Shynkaruk, O.; Horiuk, Y.; Semaniuk, N. Formation of biofilms on dairy equipment and the influence of disinfectants on them. East. Eur. J. Enterp. Technol. 2017, 5, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Kołwzan, B. Analiza zjawiska biofilmu—Warunki jego powstawania i funkcjonowania. Ochr. Sr. 2011, 33, 3–14. [Google Scholar]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Marchand, S.; Block, J.D.; Jonghe, V.D.; Coorevits, A.; Heyndrickx, M.; Herman, L. Biofilm Formation in Milk Production and Processing Environments; Influence on Milk Quality and Safety. Compr. Rev. Food Sci. Food Saf. 2012, 11, 133–147. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilmann, C.; Thumm, G.; Chhatwal, G.S.; Hartleib, J.; Uekötter, A.; Peters, G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 2003, 149, 2769–2778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penadés, J.R. Bap, a Staphylococcus aureus Surface Protein Involved in Biofilm Formation Staphylococcus aureus Surface Protein Involved in Biofilm Formation. Microbiology 2001, 183, 2888–2896. [Google Scholar]
- Puah, S.M.; Tan, J.A.M.A.; Chew, C.H.; Chua, K.H. Diverse Profiles of Biofilm and Adhesion Genes in Staphylococcus aureus Food Strains Isolated from Sushi and Sashimi. J. Food Sci. 2018, 83, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Chee, C.F.; Richter, K.; Thomas, N.; Rahman, N.A.; Nathan, S. Suppression of Staphylococcus aureus biofilm formation and virulence by a benzimidazole derivative, UM-C162. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Bowden, M.G.; Chen, W.; Singvall, J.; Xu, Y.; Peacock, S.J.; Valtulina, V.; Speziale, P.; Höök, M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 2005, 151, 1453–1464. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Singh, K.P.; Jain, A. Medical significance and management of staphylococcal biofilm. FEMS Immunol. Med. Microbiol. 2010, 58, 147–160. [Google Scholar] [CrossRef]
- Jarosińska, A.; Barłowska, J.; Wolanciuk, A.; Pastuszka, R.; Barłowska, K. Skład chemiczny i jakość mikrobiologiczna mleka towarowego dostarczanego do 5 mleczarni z regionu lubelskiego, z uwzględnieniem sezonu skupu. Rocz. Naukowe Pol. Tow. Zootech. 2014, 10, 47–56. [Google Scholar]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [Green Version]
- Van Houdt, R.; Michiels, C.W. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 2010, 109, 1117–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, K.M.; El-Razik, K.A.A.; Marie, H.S.H.; Arafa, A. Relevance of biofilm formation and virulence of different species of coagulase-negative staphylococci to public health. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Simojoki, H.; Hyvönen, P.; Ferrer, C.P.; Taponen, S.; Pyörälä, S. Is the biofilm formation and slime producing ability of coagulase-negative staphylococci associated with the persistence and severity of intramammary infection? Vet. Microbiol. 2012, 158, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.J. Bovine mastitis: An evolving disease. Vet. J. 2002, 164, 116–128. [Google Scholar] [CrossRef]
- Tremblay, Y.D.N.; Lamarche, D.; Chever, P.; Haine, D.; Messier, S.; Jacques, M. Characterization of the ability of coagulase-negative staphylococci isolated from the milk of Canadian farms to form biofilms. J. Dairy Sci. 2013, 96, 234–246. [Google Scholar] [CrossRef] [Green Version]
- Zell, C.; Resch, M.; Rosenstein, R.; Albrecht, T.; Hertel, C.; Götz, F. Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. Int. J. Food Microbiol. 2008, 127, 246–251. [Google Scholar] [CrossRef]
- Zaatout, N.; Ayachi, A.; Kecha, M.; Kadlec, K. Identification of staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J. Appl. Microbiol. 2019, 127, 1305–1314. [Google Scholar] [CrossRef]
- Kot, B.; Binek, T.; Piechota, M.; Wolska, K.M.; Zdunek, E.; Platkowska, K. Virulence factors and ability of staphylococci from bovine milk and the cowshed environment to biofilm formation. Pol. J. Vet. Sci. 2013, 16, 639–645. [Google Scholar] [CrossRef] [Green Version]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Bonaventura, G.D.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [Green Version]
- Koreňová, J.; Lopašovská, J.; Kuchta, T. Comparison of three microtitre plate-based methods for quantification of biofilm formation ability of bacteria contaminating food technologies. J. Food Nutr. Res. 2008, 47, 100–104. [Google Scholar]
- Cruzado-Bravo, M.L.M.; Silva, N.C.C.; Rodrigues, M.X.; Silva, G.O.E.; Porto, E.; Sturion, G.L. Phenotypic and genotypic characterization of Staphylococcus spp. isolated from mastitis milk and cheese processing: Study of adherence and biofilm formation. Food Res. Int. 2019, 122, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Darwish, S.F.; Asfour, H.A.E. Investigation of biofilm forming ability in staphylococci causing bovine mastitis using phenotypic and genotypic assays. Sci. World J. 2013, 2013, 378492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, P.D.C.; Ferreira, L.M.; Filho, A.N.; Zafalon, L.F.; Vicente, H.I.G.; de Souza, V. Comparison of methods for the detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Braz. J. Microbiol. 2013, 44, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, M.; Bexiga, R.; Nunes, S.F.; Carneiro, C.; Cavaco, L.M.; Bernardo, F.; Vilela, C.L. Biofilm-forming ability profiling of Staphylococcus aureus and Staphylococcus epidermidis mastitis isolates. Vet. Microbiol. 2006, 118, 133–140. [Google Scholar] [CrossRef]
- Rachmawati, D.; Alimsardjono, L. The correlation between icaA and icaD genes with biofilm formation Staphylococcus epidermidis in vitro. Folia Medica Indones. 2019, 55, 251–259. [Google Scholar] [CrossRef]
- Srednik, M.E.; Tremblay, Y.D.N.; Labrie, J.; Archambault, M.; Jacques, M.; Cirelli, A.F.; Gentilini, E.R. Biofilm formation and antimicrobial resistance genes of coagulase-negative staphylococci isolated from cows with mastitis in Argentina. FEMS Microbiol. Lett. 2017, 364, 1–8. [Google Scholar] [CrossRef]
- Dhanawade, N.B.; Kalorey, D.R.; Srinivasan, R.; Barbuddhe, S.B.; Kurkure, N.V. Detection of intercellular adhesion genes and biofilm production in Staphylococcus aureus isolated from bovine subclinical mastitis. Vet. Res. Commun. 2010, 34, 81–89. [Google Scholar] [CrossRef]
- Vasudevan, P.; Nair, M.K.M.; Annamalai, T.; Venkitanarayanan, K.S. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet. Microbiol. 2003, 92, 179–185. [Google Scholar] [CrossRef]
- Vancraeynest, D.; Hermans, K.; Haesebrouck, F. Genotypic and phenotypic screening of high and low virulence Staphylococcus aureus isolates from rabbits for biofilm formation and MSCRAMMs. Vet. Microbiol. 2004, 103, 241–247. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, D.; Shi, L.; Cai, R.; Li, C.; Yan, H. Association between agr type, virulence factors, biofilm formation and antibiotic resistance of Staphylococcus aureus isolates from pork production. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pereyra, E.A.L.; Picech, F.; Renna, M.S.; Baravalle, C.; Andreotti, C.S.; Russi, R.; Calvinho, L.F.; Diez, C.; Dallard, B.E. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet. Microbiol. 2016, 183, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, A.; Findik, A.; Onuk, E.E.; Savasan, S. Detection of methicillin resistance and slime factor production of Staphylococcus aureus in bovine mastitis. Braz. J. Microbiol. 2009, 40, 254–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woroszyło, M.; Pendrak, K.; Ciecholewska, D.; Padzik, N.; Szewczuk, M.; Karakulska, J. Investigation of Biofilm Formation Ability of Coagulase-Negative Staphylococci Isolated from Ready-to-Eat Meat. Acta Sci. Pol. Zootech. 2018, 17, 27–36. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, S.; Lou, X.; Cheng, S.; Liu, X.; Zheng, W.; Zheng, Z.; Wang, H. Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. Microbiology open 2020, 9, e00946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirmusaoglu, S. Staphylococcal Biofilms: Pathogenicity, Mechanism and Regulation of Biofilm Formation by Quorum-Sensing System and Antibiotic Resistance Mechanisms of Biofilm-Embedded Microorganisms. In Microbial Biofilms-Importance and Application; InTech: Vienna, Austria, 2016; pp. 189–209. [Google Scholar]
- Khoramian, B.; Jabalameli, F.; Niasari-Naslaji, A.; Taherikalani, M.; Emaneini, M. Comparison of virulence factors and biofilm formation among Staphylococcus aureus strains isolated from human and bovine infections. Microb. Pathog. 2015, 88, 73–77. [Google Scholar] [CrossRef]
- Vautor, E.; Abadie, G.; Pont, A.; Thiery, R. Evaluation of the presence of the bap gene in Staphylococcus aureus isolates recovered from human and animals species. Vet. Microbiol. 2008, 127, 407–411. [Google Scholar] [CrossRef] [Green Version]
- Tormo, M.Á.; Knecht, E.; Götz, F.; Lasa, I.; Penadés, J.R. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: Evidence of horizontal gene transfer? Microbiology 2005, 151, 2465–2475. [Google Scholar] [CrossRef] [Green Version]
- Martins, K.B.; Faccioli, P.Y.; Bonesso, M.F.; Fernandes, S.; Oliveira, A.A.; Dantas, A.; Zafalon, L.F.; Cunha, M.D.L.R.S. Characteristics of resistance and virulence factors in different species of coagulase-negative staphylococci isolated from milk of healthy sheep and animals with subclinical mastitis. J. Dairy Sci. 2017, 100, 2184–2195. [Google Scholar] [CrossRef] [Green Version]
- Malinowski, E.; Kłossowska, A.; Zastempowska, E. Virulence factors in coagulase-negative staphylococci isolated from cows with subclinical mastitis. Bull. Vet. Inst. Pulawy 2011, 55, 681–684. [Google Scholar]
- Mathur, T.; Singhal, S.; Khan, S.; Upadhyay, D.J.; Fatma, T.; Rattan, A. Detection of biofilm formation among the clinical isolates of staphylococci: An evaluation of three different screening methods. Indian J. Med. Microbiol. 2006, 24, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćircović, I.; Ruzicka, F. Quantification of biofilm in microtiter plates. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
| Strains | Biofilm Production (MTP Method) | Slime Production (CRA Method) | |||
|---|---|---|---|---|---|
| Strong | Moderate | Weak | Positive | Negative | |
| S. haemolyticus (n = 11) | 8 (72.7%) | 1 (9.1%) | 2 (18.2%) | 4 (36.4%) | 7 (63.6%) |
| S. simulans (n = 8) | 7 (87.5%) | - | 1 (12.5%) | 7 (87.5%) | 1 (12.5%) |
| S. epidermidis (n = 6) | 5 (83.3%) | 1 (16.7%) | - | 4 (66.7%) | 2 (33.3%) |
| S. warneri (n = 5) | 4 (80%) | - | 1 (20%) | 2 (40%) | 3 (60%) |
| S. chromogenes (n = 4) | 4 (100%) | - | - | 2 (50%) | 2 (50%) |
| S. hominis (n = 3) | 3 (100%) | - | - | 1 (33.3%) | 2 (66.7%) |
| S. sciuri (n = 2) | 2 (100%) | - | - | 1 (50%) | 1 (50%) |
| S. saprophyticus (n = 1) | 1 (100%) | - | - | 1 (100%) | - |
| S. capitis (n = 1) | 1 (100%) | - | - | - | 1 (100%) |
| S. xylosus (n = 1) | 1 (100%) | - | - | 1 (100%) | - |
| Total CoNS (n = 42) | 36 (85.7%) | 2 (4.8%) | 4 (9.5%) | 23 (54.8%) | 19 (45.2%) |
| Slime producing CoNS (n = 23) | 20 (87%) | 1 (4.3%) | 2 (8.7%) | - | - |
| No slime-producing CoNS (n = 19) | 16 (84.2%) | 1 (5.3%) | 2 (10.5%) | - | - |
| S. aureus (n = 12) | 7 (58.3%) | 1 (8.3%) | 4 (33.3%) | 5 (41.7%) | 7 (58.3%) |
| Slime producing S. aureus (n = 5) | 4 (80%) | 0 | 1 (20%) | - | - |
| No slime-producing S. aureus (n = 7) | 3 (42.9%) | 1 (14.2%) | 3 (42.9%) | - | - |
| All staphylococci (n = 54) | 43 (79.6%) | 3 (5.6%) | 8 (14.8%) | 28 (51.9%) | 26 (48.1%) |
| Biofilm-Associated Gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strains | icaA | icaD | bap | eno | aap | fbe | bhp | embP | atlE |
| S. haemolyticus (n = 11) | 5 (45.5%) | 2 (18.2%) | 3 (27.3%) | 7 (63.6%) | 3 (27.3%) | 2 18,2%) | - | - | - |
| S. simulans (n = 8) | 4 (50%) | 2 (25%) | 1 (12.5%) | 2 (25%) | 3 (37.5%) | - | - | 2 (25%) | 5 (62.5%) |
| S. epidermidis (n = 6) | - | 2 (33.3%) | 3 (50%) | 4 (66.7%) | 5 (60%) | 1 (16.7%) | - | 4 (66.7%) | 4 (66.7%) |
| S. warneri (n = 5) | 1 (20%) | 1 (20%) | 1 (20%) | 3 (60%) | - | - | - | - | - |
| S. chromogenes (n = 4) | 1 (25%) | - | - | 2 (50%) | - | - | - | - | - |
| S. hominis (n = 3) | 1 (33.3%) | - | - | - | - | - | - | - | 1 (33.3%) |
| S. sciuri (n = 2) | 1 (50%) | - | 2 (100%) | 2 (100%) | 1 (50%) | - | - | 1 (50%) | - |
| S. sapropythicus (n = 1) | - | 1 (100%) | - | - | - | - | - | - | - |
| S. capitis (n = 1) | - | 1 (100%) | - | - | - | - | - | - | - |
| S. xylosus (n = 1) | - | - | - | - | - | - | - | - | - |
| Total CoNS (n = 42) | 12 (31.0%) | 9 (21.4%) | 10 (23.8%) | 20 (47.6%) | 12 (28.6%) | 3 (7.1%) | - | 7 (16.7%) | 10 (23.8%) |
| S. aureus (n = 12) | - | 12 (100%) | 3 (25%) | 7 (58.3%) | NA | NA | NA | NA | NA |
| All staphylococci (n = 54) | 13 (24.1%) | 21 (38.9%) | 13 (24.1%) | 27 (50.0%) | - | - | - | - | - |
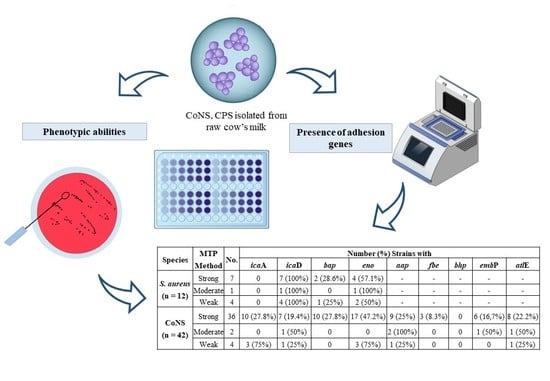
| Species | MTP Method | No. | Number (%) Strains with | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| icaA | icaD | bap | eno | aap | fbe | bhp | embP | atlE | |||
| S. aureus (n = 12) | Strong | 7 | 0 | 7 (100%) | 2 (28.6%) | 4 (57.1%) | - | - | - | - | - |
| Moderate | 1 | 0 | 1 (100%) | 0 | 1 (100%) | - | - | - | - | - | |
| Weak | 4 | 0 | 4 (100%) | 1 (25%) | 2 (50%) | - | - | - | - | - | |
| CoNS (n = 42) | Strong | 36 | 10 (27.8%) | 7 (19.4%) | 10 (27.8%) | 17 (47.2%) | 9 (25%) | 3 (8.3%) | 0 | 6 (16,7%) | 8 (22.2%) |
| Moderate | 2 | 0 | 1 (50%) | 0 | 0 | 2 (100%) | 0 | 0 | 1 (50%) | 1 (50%) | |
| Weak | 4 | 3 (75%) | 1 (25%) | 0 | 3 (75%) | 1 (25%) | 0 | 0 | 0 | 1 (25%) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajewska, J.; Chajęcka-Wierzchowska, W. Biofilm Formation Ability and Presence of Adhesion Genes among Coagulase-Negative and Coagulase-Positive Staphylococci Isolates from Raw Cow’s Milk. Pathogens 2020, 9, 654. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9080654
Gajewska J, Chajęcka-Wierzchowska W. Biofilm Formation Ability and Presence of Adhesion Genes among Coagulase-Negative and Coagulase-Positive Staphylococci Isolates from Raw Cow’s Milk. Pathogens. 2020; 9(8):654. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9080654
Chicago/Turabian StyleGajewska, Joanna, and Wioleta Chajęcka-Wierzchowska. 2020. "Biofilm Formation Ability and Presence of Adhesion Genes among Coagulase-Negative and Coagulase-Positive Staphylococci Isolates from Raw Cow’s Milk" Pathogens 9, no. 8: 654. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9080654