Bioremediation of Petroleum-Contaminated Soils with Biosurfactant-Producing Degraders Isolated from the Native Desert Soils
Abstract
:1. Introduction
2. Materials and Methods
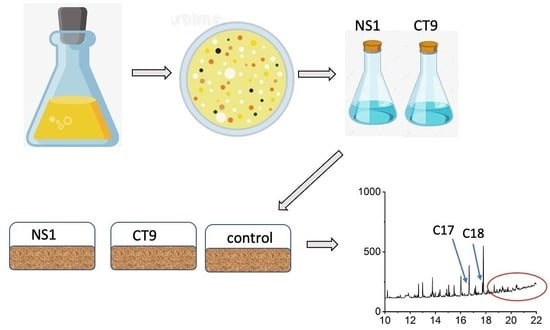
2.1. Enrichment and Isolation of Biosurfactant-Producing Strains
2.2. Characterization of the Isolates by 16S rRNA Sequencing and GEN III MicroPlate
2.3. Screening of Biosurfactant Production Activity
2.4. Preparation of Inoculum and Soil Batch Incubation
2.5. Bioaugmentation Experiment
2.6. DNA Extraction, Real-Time Polymerase Chain Reaction (qPCR), Illumina Sequencing, and Microbial Community Analyses
2.6.1. DNA Extraction and qPCR
2.6.2. Amplicon Sequencing
2.6.3. Bioinformatics
2.7. Soil Hydrophobicity and Hydrocarbon Analysis
3. Results
3.1. Biosurfactants Screening
3.2. Isolates Characterization
3.3. Bioaugmentation Experiment
3.3.1. Microbial Community Change
3.3.2. Functional Gene Abundance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, Y.; Wei, Y.; Qian, H.; Fang, Y. Analysis of the groundwater and soil pollution by oil leakage. Procedia Environ. Sci. 2011, 11, 939–944. [Google Scholar] [CrossRef]
- Hewelke, E.; Oktaba, L.; Gozdowski, D.; Kondras, M.; Olejniczak, I.; Górska, E.B. Intensity and persistence of soil water repellency in pine forest soil in a temperate continental climate under drought conditions. Water 2018, 10, 1121. [Google Scholar] [CrossRef]
- Roy, A.; Dutta, A.; Pal, S.; Gupta, A.; Sarkar, J.; Chatterjee, A.; Saha, A.; Sarkar, P.; Sar, P.; Kazy, S.K. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour. Technol. 2018, 253, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J. The basis of the hydrophobic effect. Biophys. Chem. 2002, 100, 193–203. [Google Scholar] [CrossRef]
- Weissenfels, W.D.; Klewer, H.J.; Langhoff, J. Adsorption of polycyclic aromatic hydrocarbons (PAHs) by soil particles: Influence on biodegradability and biotoxicity. Appl. Microbiol. Biotechnol. 1992, 36, 689–696. [Google Scholar] [CrossRef]
- Banet, G.; Turaani, A.K.; Farber, R.; Armoza-Zvuloni, R.; Rotem, N.; Stavi, I.; Cahan, R. The effects of biostimulation and bioaugmentation on crude oil biodegradation in two adjacent terrestrial oil spills of different age, in a hyper-arid region. J. Environ. Manag. 2021, 286, 112248. [Google Scholar] [CrossRef]
- Li, Z.; Ronen, Z.; Gelman, F.; Crouvi, O.; Arye, G.; Rosenzweig, R. Reclamation of oil-induced soil hydrophobicity in the hyper-arid Evrona Nature Reserve, southern Israel. Pedosphere 2021, 31, 892–902. [Google Scholar] [CrossRef]
- Gordon, G.; Stavi, I.; Shavit, U.; Rosenzweig, R. Oil spill effects on soil hydrophobicity and related properties in a hyper-arid region. Geoderma 2018, 312, 114–120. [Google Scholar] [CrossRef]
- Stavi, I.; Rosenzweig, R. Tillage effect on hydrophobicity and hydrological properties of oil-contaminated sediments in a hyper-arid region. Arid L. Res. Manag. 2020, 34, 26–35. [Google Scholar] [CrossRef]
- Jain, P.K.; Gupta, V.K.; Gaur, R.K.; Lowry, M.; Jaroli, D.P.; Chauhan, U.K. Bioremediation of petroleum oil contaminated soil and water. Res. J. Environ. Toxicol. 2011, 5, 1. [Google Scholar]
- Lim, M.W.; Von Lau, E.; Poh, P.E. A comprehensive guide of remediation technologies for oil contaminated soil—Present works and future directions. Mar. Pollut. Bull. 2016, 109, 14–45. [Google Scholar] [CrossRef]
- Eze, M.O.; Thiel, V.; Hose, G.C.; George, S.C.; Daniel, R. Bacteria-plant interactions synergistically enhance biodegradation of diesel fuel hydrocarbons. Commun. Earth Environ. 2022, 3, 192. [Google Scholar] [CrossRef]
- Crisafi, F.; Genovese, M.; Smedile, F.; Russo, D.; Catalfamo, M.; Yakimov, M.; Giuliano, L.; Denaro, R. Bioremediation technologies for polluted seawater sampled after an oil-spill in Taranto Gulf (Italy): A comparison of biostimulation, bioaugmentation and use of a washing agent in microcosm studies. Mar. Pollut. Bull. 2016, 106, 119–126. [Google Scholar] [CrossRef]
- Menezes Bento, F.; de Oliveira Camargo, F.A.; Okeke, B.C.; Frankenberger, W.T. Diversity of biosurfactant producing microorganisms isolated from soils contaminated with diesel oil. Microbiol. Res. 2005, 160, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.L.; Letey, J.; Wu, L. The Influence of Two Surfactants on Infiltration into a Water-Repellent Soil. Soil Sci. Soc. Am. J. 2002, 66, 361–367. [Google Scholar] [CrossRef]
- Banat, I.M. Biosurfactants production and possible uses in microbial enhanced oil recovery and oil pollution remediation: A review. Bioresour. Technol. 1995, 51, 1–12. [Google Scholar] [CrossRef]
- Ibrahim, M.; Ijah, U.J.J.; Manga, S.B.; Bilbis, L.S.; Umar, S. Production and partial characterization of biosurfactant produced by crude oil degrading bacteria. Int. Biodeterior. Biodegrad. 2013, 81, 28–34. [Google Scholar] [CrossRef]
- Patowary, R.; Patowary, K.; Kalita, M.C.; Deka, S. Application of biosurfactant for enhancement of bioremediation process of crude oil contaminated soil. Int. Biodeterior. Biodegrad. 2018, 129, 50–60. [Google Scholar] [CrossRef]
- Aparna, A. Effect of addition of biosurfactant produced by Pseudomonas sps. on biodegradation of crude oil. In Proceedings of the 2nd International Conference on Environmental Science and Technology, Singapore, 26–28 February 2011; Volume 6, pp. 71–75. [Google Scholar]
- Thavasi, R.; Jayalakshmi, S.; Banat, I.M. Application of biosurfactant produced from peanut oil cake by Lactobacillus delbrueckii in biodegradation of crude oil. Bioresour. Technol. 2011, 102, 3366–3372. [Google Scholar] [CrossRef]
- Kang, S.-W.; Kim, Y.-B.; Shin, J.-D.; Kim, E.-K. Enhanced biodegradation of hydrocarbons in soil by microbial biosurfactant, Sophorolipid. Appl. Biochem. Biotechnol. 2010, 160, 780–790. [Google Scholar] [CrossRef]
- Rahman, K.S.M.; Banat, I.M.; Thahira, J.; Thayumanavan, T.; Lakshmanaperumalsamy, P. Bioremediation of gasoline contaminated soil by a bacterial consortium amended with poultry litter, coir pith and rhamnolipid biosurfactant. Bioresour. Technol. 2002, 81, 25–32. [Google Scholar] [CrossRef]
- Mnif, I.; Mnif, S.; Sahnoun, R.; Maktouf, S.; Ayedi, Y.; Ellouze-Chaabouni, S.; Ghribi, D. Biodegradation of diesel oil by a novel microbial consortium: Comparison between co-inoculation with biosurfactant-producing strain and exogenously added biosurfactants. Environ. Sci. Pollut. Res. 2015, 22, 14852–14861. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using Next-Generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure–function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Cooper, D.G.; Goldenberg, B.G. Surface-active agents from two bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef]
- Sidkey, N.; Mohamed, H.; Elkhouly, H. Evaluation of different dcreening methods for biosurfactant producers isolated from contaminated Egyptian samples grown on industrial olive oil processing waste. Br. Microbiol. Res. J. 2016, 17, 1–19. [Google Scholar] [CrossRef]
- Joy, S.; Rahman, P.K.S.M.; Sharma, S. Biosurfactant production and concomitant hydrocarbon degradation potentials of bacteria isolated from extreme and hydrocarbon contaminated environments. Chem. Eng. J. 2017, 317, 232–241. [Google Scholar] [CrossRef]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, I.; Wagner, F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 1991, 5, 265–268. [Google Scholar] [CrossRef]
- Marchant, R.; Sharkey, F.H.; Banat, I.M.; Rahman, T.J.; Perfumo, A. The degradation of n-hexadecane in soil by thermophilic geobacilli. FEMS Microbiol. Ecol. 2006, 56, 44–54. [Google Scholar] [CrossRef]
- Zhao, F.; Shi, R.; Ma, F.; Han, S.; Zhang, Y. Oxygen effects on rhamnolipids production by Pseudomonas aeruginosa. Microb. Cell Fact. 2018, 17, 39. [Google Scholar] [CrossRef]
- Ionescu, D.; Overholt, W.A.; Lynch, M.D.J.; Neufeld, J.D.; Naqib, A.; Green, S.J. Microbial Community Analysis Using High-Throughput Amplicon Sequencing. In Manual of Environmental Microbiology; Wiley: Hoboken, NJ, USA, 2016; pp. 2–4. ISBN 9781683670742. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, X.; Mansoldo, F.R.P.; An, J.; Kou, Y.; Zhang, X.; Wang, J.; Zeng, J.; Vermelho, A.B.; Yao, M. Microbial habitat specificity largely affects microbial co-occurrence patterns and functional profiles in wetland soils. Geoderma 2022, 418, 115866. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome R Package. 2017. Available online: https://www.bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 1 August 2022).
- Letey, J. Measurement of Contact Angle, Water Drop Penetration Time, and Critical Surface Tension. In Proceedings of the Symposium on Water Repellant Soils, Riverside, CA, USA, 6–10 May 1968. [Google Scholar]
- Sarand, I.; Haario, H.; Jørgensen, K.S.; Romantschuk, M. Effect of inoculation of a TOL plasmid containing mycorrhizosphere bacterium on development of Scots pine seedlings, their mycorrhizosphere and the microbial flora in m-toluate-amended soil. FEMS Microbiol. Ecol. 2000, 31, 127–141. [Google Scholar] [CrossRef]
- Wang, Y.; Kou, S.; Jiang, Q.; Xu, B.; Liu, X.; Xiao, J.; Tian, Y.; Zhou, C.; Zhang, C.; Xiao, M. Factors affecting transfer of degradative plasmids between bacteria in soils. Appl. Soil Ecol. 2014, 84, 254–261. [Google Scholar] [CrossRef]
- Satpute, S.K.; Bhawsar, B.D.; Dhakephalkar, P.K.; Chopade, B.A. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Indian J. Mar. Sci. 2008, 37, 243–250. [Google Scholar]
- Monteiro, S.A.; Sassaki, G.L.; de Souza, L.M.; Meira, J.A.; de Araújo, J.M.; Mitchell, D.A.; Ramos, L.P.; Krieger, N. Molecular and structural characterization of the biosurfactant produced by Pseudomonas aeruginosa DAUPE 614. Chem. Phys. Lipids 2007, 147, 1–13. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, D.; Zhu, C.; Lundaa, T.; Scherr, K.E. Isolation and identification of biosurfactant producing and crude oil degrading Pseudomonas aeruginosa strains. Chem. Eng. J. 2012, 209, 138–146. [Google Scholar] [CrossRef]
- Mulligan, C.N. Environmental applications for biosurfactants. Environ. Pollut. 2005, 133, 183–198. [Google Scholar] [CrossRef]
- Kamyabi, A.; Nouri, H.; Moghimi, H. Synergistic Effect of Sarocladium sp. and Cryptococcus sp. Co-Culture on Crude Oil Biodegradation and Biosurfactant Production. Appl. Biochem. Biotechnol. 2017, 182, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Patowary, K.; Saikia, R.R.; Kalita, M.C.; Deka, S. Degradation of polyaromatic hydrocarbons employing biosurfactant-producing Bacillus pumilus KS2. Ann. Microbiol. 2015, 65, 225–234. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Y.; Zang, T.; Wei, J.; Wu, H.; Wei, C.; Qiu, G.; Li, F. A biosurfactant-producing Pseudomonas aeruginosa S5 isolated from coking wastewater and its application for bioremediation of polycyclic aromatic hydrocarbons. Bioresour. Technol. 2019, 281, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Bey Baba Hamed, M.; El-Amine Abi Ayad, S.-M.; Faiza, B.M. Hydrocarbon-Degrading Bacterial Strain Pseudomonas mendocina Newly Isolated from Marine Sediments and Seawater of Oran Harbor (Algerian Coast). Arch. Ecotoxicol. 2020, 2, 22–29. [Google Scholar] [CrossRef]
- Saruni, N.H.; Abdul Razak, S.; Habib, S.; Ahmad, S.A.; Alias, S.A.; Wan Johari, W.L.; Smykla, J.; Yasid, N.A. Comparative Screening Methods for the Detection of Biosurfactant-Producing Capability of Antarctic Hydrocarbon-degrading Pseudomonas sp. J. Environ. Microbiol. Toxicol. 2019, 7, 44–47. [Google Scholar] [CrossRef]
- Tarhriz, V.; Nouioui, I.; Spröer, C.; Verbarg, S.; Ebrahimi, V.; Cortés-Albayay, C.; Schumann, P.; Hejazi, M.A.; Klenk, H.-P.; Hejazi, M.S. Pseudomonas khazarica sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from Khazar Sea sediments. Antonie Van Leeuwenhoek 2020, 113, 521–532. [Google Scholar] [CrossRef]
- Wu, T.; Xu, J.; Xie, W.; Yao, Z.; Yang, H.; Sun, C.; Li, X. Pseudomonas aeruginosa L10: A hydrocarbon-degrading, biosurfactant-producing, and plant-growth-promoting endophytic bacterium isolated from a Reed (Phragmites australis). Front. Microbiol. 2018, 9, 1087. [Google Scholar] [CrossRef]
- Muthukrishnan, T.; Abed, R.M.M. Effects of Irrigation on Alkane Biodegradation of Oil-Contaminated Desert Soils. Environ. Process. 2018, 5, 631–648. [Google Scholar] [CrossRef]
- Marín-García, D.C.; Adams, R.H.; Hernández-Barajas, R. Effect of crude petroleum on water repellency in a clayey alluvial soil. Int. J. Environ. Sci. Technol. 2016, 13, 55–64. [Google Scholar] [CrossRef]
- Adams, R.H.; Guzmán Osorio, F.J.; Zavala Cruz, J. Water repellency in oil contaminated sandy and clayey soils. Int. J. Environ. Sci. Technol. 2008, 5, 445–454. [Google Scholar] [CrossRef]







| Isolate Code | Oil Spreading | E24 Index | Surface Tension (mN/m) a | Surface Tension (mN/m) b | CTAB Halo Area (mm2) |
|---|---|---|---|---|---|
| NS1 | + | 48.2% ± 0.5% (+) | 41.86 ± 0.03 (++) | 27.70 ± 0.04 (++++) | 178 ± 15.59 |
| NS2 | + | 44.4% ± 0.9% (+) | 43.70 ± 0.04 (++) | 28.92 ± 0.04 (++++) | 186.83 ± 7.94 |
| NS3 | + | 51.9% ± 0.4% (++) | 44.11 ± 0.07 (++) | 27.56 ± 0.08 (++++) | 196.17 ± 14.00 |
| NS4 | + | 42.6% ± 0.3% (+) | 44.50 ± 0.04 (++) | 27.63 ± 0.03 (++++) | 200.75 ± 8.23 |
| NS5 | + | 48.2% ± 0.5% (+) | 43.19 ± 0.08 (++) | 27.67 ± 0.04 (++++) | 186.83 ± 7.94 |
| NS6 | + | 48.2% ± 0.7% (+) | 44.10 ± 0.03 (++) | 28.82 ± 0.03 (++++) | 205.5 ± 8.23 |
| NS8 | + | 51.9% ± 0.2% (++) | 41.81 ± 0.05 (++) | 27.65 ± 0.02 (++++) | 196.94 ± 1.62 |
| CT7 | - | - | 56.40 ± 0.09 (+) | / | / |
| CT9 | - | - | 56.76 ± 0.02 (+) | / | / |
| CT10 | - | - | 55.51 ± 0.03 (+) | / | / |
| CT11 | - | - | 56.15 ± 0.04 (+) | / | / |
| CT12 | - | - | 55.39 ± 0.07 (+) | / | / |
| CT14 | - | - | 54.22 ± 0.05′ (+) | / | / |
| PAO1 | + | 63% ± 0.9% (+++) | 40.03 ± 0.05 (++) | 29.61 ± 0.03 (++++) | 227.1 ± 3.49 |
| Neg | - | - | 68.13 ± 0.03 (-) | 64.06 ± 0.07 (-) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Rosenzweig, R.; Chen, F.; Qin, J.; Li, T.; Han, J.; Istvan, P.; Diaz-Reck, D.; Gelman, F.; Arye, G.; et al. Bioremediation of Petroleum-Contaminated Soils with Biosurfactant-Producing Degraders Isolated from the Native Desert Soils. Microorganisms 2022, 10, 2267. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10112267
Li Z, Rosenzweig R, Chen F, Qin J, Li T, Han J, Istvan P, Diaz-Reck D, Gelman F, Arye G, et al. Bioremediation of Petroleum-Contaminated Soils with Biosurfactant-Producing Degraders Isolated from the Native Desert Soils. Microorganisms. 2022; 10(11):2267. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10112267
Chicago/Turabian StyleLi, Zheng, Ravid Rosenzweig, Fengxian Chen, Ji Qin, Tianyi Li, Jincheng Han, Paula Istvan, Damiana Diaz-Reck, Faina Gelman, Gilboa Arye, and et al. 2022. "Bioremediation of Petroleum-Contaminated Soils with Biosurfactant-Producing Degraders Isolated from the Native Desert Soils" Microorganisms 10, no. 11: 2267. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10112267