Antibiotic Resistance, Virulence Factors, Phenotyping, and Genotyping of E. coli Isolated from the Feces of Healthy Subjects
Abstract
:1. Introduction
2. Results
2.1. Quantitation, Phyloptyping, and Genotyping
2.2. Virulence Factors
2.3. Other Genetic Determinants Affecting Fitness and Pathogenicity
2.4. Antibiotic Susceptibility
2.5. Conjugation
2.6. Production of Curli and Cellulose and Biofilm Formation
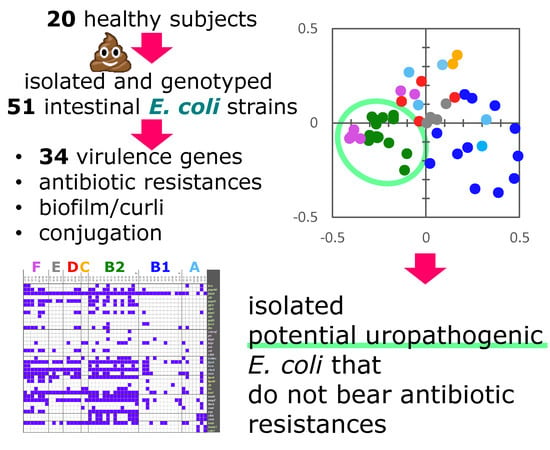
2.7. Co-Occurrence and Principal Coordinate Analysis (PCoA) Analysis of Genetic and Functional Features
3. Discussion
4. Materials and Methods
4.1. Isolation and Enumeration and of E. coli
4.2. Phyloptyping and PFGE Genotyping
4.3. Virulence Genotyping
4.4. Detection of the Genes traD, colE7, immE7, and of pks Island
4.5. Antibiotic Susceptibility
4.6. Biofilm and Phenotype Assays
4.7. Solid Mating Conjugation Experiments
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Russo, T.A.; Johnson, J.R. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J. Infect. Dis. 2000, 181, 1753–1754. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Leimbach, A.; Hacker, J.; Dobrindt, U. E. coli as an all-rounder: The thin line between commensalism and pathogenicity. Curr. Top Microbiol. Immunol. 2013, 358, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Searle, L.J.; Méric, G.; Porcelli, I.; Sheppard, S.K.; Lucchini, S. Variation in siderophore biosynthetic gene distribution and production across environmental and faecal populations of Escherichia coli. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Welch, R.A. Uropathogenic Escherichia coli-Associated Exotoxins. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Kaas, R.S.; Friis, C.; Ussery, D.W.; Aarestrup, F.M. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genom. 2012, 13, 577. [Google Scholar] [CrossRef]
- Koraimann, G. Spread and persistence of virulence and antibiotic resistance genes: A ride on the F plasmid conjugation module. EcoSal Plus 2018, 8, 1. [Google Scholar] [CrossRef]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef]
- Kudinha, T.; Johnson, J.R.; Andrew, S.D.; Kong, F.; Anderson, P.; Gilbert, G.L. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin. Microbiol. Infect. 2013, 19. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Han, D.S.; Jo, S.V.; Lee, A.R.; Park, C.H.; Eun, C.S.; Lee, Y. Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: Potential impact on clinical outcomes. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Commensal Escherichia coli of healthy humans: A reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 2010, 59, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Mathai, D.; Kumar, V.A.; Paul, B.; Sugumar, M.; John, K.R.; Manoharan, A.; Kesavan, L.M. Fecal carriage rates of extended-spectrum β-lactamase-producing Escherichia coli among antibiotic naive healthy human volunteers. Microb. Drug Resist. 2015, 21, 59–64. [Google Scholar] [CrossRef] [PubMed]
- González, D.; Gallagher, E.; Zúñiga, T.; Leiva, J.; Vitas, A.I. Prevalence and characterization of β-lactamase-producing Enterobacteriaceae in healthy human carriers. Int. Microbio. 2019, in press. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Wassenaar, T.M. E. coli and colorectal cancer: A complex relationship that deserves a critical mindset. Crit. Rev. Microbiol. 2018, 44, 619–632. [Google Scholar] [CrossRef]
- Frost, L.S.; Ippen-Ihler, K.; Skurray, R.A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 1994, 58, 162–210. [Google Scholar]
- Chak, K.F.; Kuo, W.S.; Lu, F.M.; James, R. Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol. 1991, 137, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Raimondi, S.; Amaretti, A.; Gentilomi, G.; Bonvicini, F.; Rossi, M. Antibiotic resistance, virulence factors, phenotyping, and genotyping of Enterobacteriaceae (other than E. coli) isolated from the feces of healthy subjects. Unpublished; manuscript in preparation.
- Nowrouzian, F.L.; Wold, A.E.; Adlerberth, I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis. 2005, 191, 1078–1083. [Google Scholar] [CrossRef]
- Ostblom, A.; Adlerberth, I.; Wold, A.E.; Nowrouzian, F.L. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants’ commensal microbiotas. Appl. Environ. Microbiol. 2011, 77, 2303–2308. [Google Scholar] [CrossRef]
- Robins-Browne, R.M.; Holt, K.E.; Ingle, D.J.; Hocking, D.M.; Yang, J.; Tauschek, M. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front. Cell. Infect. Microbiol. 2016, 6, 141. [Google Scholar] [CrossRef]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 5792. [Google Scholar] [CrossRef] [Green Version]
- Sahuquillo-Arce, J.M.; Selva, M.; Perpiñán, H.; Gobernado, M.; Armero, C.; López-Quílez, A.; González, F.; Vanaclocha, H. Antimicrobial resistance in more than 100,000 Escherichia coli isolates according to culture site and patient age, gender, and location. Antimicrob. Agents Chemother. 2011, 55, 1222–1228. [Google Scholar] [CrossRef]
- Cole, B.K.; Ilikj, M.; McCloskey, C.B.; Chavez-Bueno, S. Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Morley, V.J.; Woods, R.J.; Read, A.F. Bystander selection for antimicrobial resistance: Implications for patient health. Trends in Microbiol. 2019, in press. [Google Scholar] [CrossRef]
- Dyar, O.J.; Hoa, N.Q.; Trung, N.V.; Phuc, H.D.; Larsson, M.; Chuc, N.T.; Lundborg, C.S. High prevalence of antibiotic resistance in commensal Escherichia coli among children in rural Vietnam. BMC Infect. Dis. 2012, 12, 92. [Google Scholar] [CrossRef]
- Gurnee, E.A.; Ndao, I.M.; Johnson, J.R.; Johnston, B.D.; Gonzalez, M.D.; Burnham, C.A.; Hall-Moore, C.M.; McGhee, J.E.; Mellmann, A.; Warner, B.B.; et al. Gut colonization of healthy children and their mothers with pathogenic ciprofloxacin-resistant Escherichia coli. J. Infect. Dis. 2015, 212, 1862–1868. [Google Scholar] [CrossRef]
- Bag, S.; Ghosh, T.S.; Banerjee, S.; Mehta, O.; Verma, J.; Dayal, M.; Desigamani, A.; Kumar, P.; Saha, B.; Kedia, S.; et al. insights into antimicrobial resistance traits of commensal human gut microbiota. Microb. Ecol. 2019, 77, 546–557. [Google Scholar] [CrossRef]
- Purohit, M.R.; Lindahl, L.F.; Diwan, V.; Marrone, G.; Lundborg, C.S. High levels of drug resistance in commensal E. coli in a cohort of children from rural central India. Sci. Rep. 2019, 9, 6682. [Google Scholar] [CrossRef]
- Feng, J.; Li, B.; Jiang, X.; Yang, Y.; Wells, G.F.; Zhang, T.; Li, X. Antibiotic resistome in a large-scale healthy human gut microbiota deciphered by metagenomic and network analyses. Environ. Microbiol. 2018, 20, 355–368. [Google Scholar] [CrossRef]
- Pormohammad, A.; Nasiri, M.J.; Azimi, T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis. Infect. Drug Resist. 2019, 12, 1181–1197. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Dinh, P.C.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Servin, A.L. Pathogenesis of Afa/Dr. diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 2005, 18, 264–292. [Google Scholar] [CrossRef]
- Biesecker, S.G.; Nicastro, L.K.; Wilson, R.P.; Tükel, Ç. The functional amyloid curli protects Escherichia coli against complement-mediated bactericidal activity. Biomolecules 2018, 8, 5. [Google Scholar] [CrossRef]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2018, 51, 167–176. [Google Scholar] [CrossRef]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Nougayrede, J.P.; Oswald, E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 2008, 46, 3906–3911. [Google Scholar] [CrossRef]
- Murase, K.; Martin, P.; Porcheron, G.; Houle, S.; Helloin, E.; Pénary, M.; Nougayrède, J.P.; Dozois, C.M.; Hayashi, T.; Oswald, E. HlyF produced by extraintestinal pathogenic Escherichia coli is a virulence factor that regulates outer membrane vesicle biogenesis. J. Infect. Dis. 2016, 213, 856–865. [Google Scholar] [CrossRef]
- Gophna, U.; Parket, A.; Hacker, J.; Ron, E.Z. A novel ColV plasmid encoding type IV pili. Microbiol. 2003, 149, 177–184. [Google Scholar] [CrossRef]
- Starčič Erjavec, M.; Petkovšek, Ž.; Kuznetsova, M.V.; Maslennikova, I.L.; Žgur-Bertok, D. Strain ŽP—The first bacterial conjugation-based “kill”-” anti-kill” antimicrobial system. Plasmid 2015, 82, 28–34. [Google Scholar] [CrossRef]
- Maslennikova, I.L.; Kuznetsova, M.V.; Toplak, N.; Nekrasova, I.V.; Žgur Bertok, D.; Starčič Erjavec, M. Estimation of the bacteriocin ColE7 conjugation-based “kill” - “anti-kill” antimicrobial system by real-time PCR, fluorescence staining and bioluminescence assays. Lett. Appl. Microbiol. 2018, 67, 47–53. [Google Scholar] [CrossRef]
- Wilson, L.A.; Sharp, P.M. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: Evolution and implications for ERIC-PCR. Mol. Biol. Evol. 2006, 23, 1156–1168. [Google Scholar] [CrossRef]
- Quartieri, A.; Simone, M.; Gozzoli, C.; Popovic, M.; D’Auria, G.; Amaretti, A.; Raimondi, S.; Rossi, M. Comparison of culture-dependent and independent approaches to characterize fecal bifidobacteria and lactobacilli. Anaerobe 2016, 38, 130–137. [Google Scholar] [CrossRef]
- Simone, M.; Gozzoli, C.; Quartieri, A.; Mazzola, G.; Di Gioia, D.; Amaretti, A.; Raimondi, S.; Rossi, M. The probiotic Bifidobacterium breve B632 inhibited the growth of Enterobacteriaceae within colicky infant microbiota cultures. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef]
- Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Available online: http://www.cdc.gov/pulsenet/PDF/ecoli-shigella-salmonella-pfge-protocol-508c.pdf (accessed on 19 July 2019).
- Johnson, J.R.; Porter, S.; Johnston, B.; Kuskowski, M.A.; Spurbeck, R.R.; Mobley, H.L.; Williamson, D.A. Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect. Dis. 2015, 2. [Google Scholar] [CrossRef]
- Sarshar, M.; Scribano, D.; Marazzato, M.; Ambrosi, C.; Aprea, M.R.; Aleandri, M.; Pronio, A.; Longhi, C.; Nicoletti, M.; Zagaglia, C.; et al. Genetic diversity, phylogroup distribution and virulence gene profile of pks positive Escherichia coli colonizing human intestinal polyps. Microb. Pathog. 2017, 112, 274–278. [Google Scholar] [CrossRef]
- Sicard, J.F.; Vogeleer, P.; Le Bihan, G.; Rodriguez Olivera, Y.; Beaudry, F.; Jacques, M.; Harel, J. N-Acetyl-glucosamine influences the biofilm formation of Escherichia coli. Gut Pathog. 2018, 22, 10–26. [Google Scholar] [CrossRef]
- Vogeleer, P.; Tremblay, Y.D.N.; Jubelin, G.; Jacques, M.; Harel, J. Biofilm-forming abilities of shiga toxin-producing Escherichia coli isolates associated with human infections. Appl. Environ. Microbiol. 2015, 28, 1448–1458. [Google Scholar] [CrossRef]







| E. coli Strain | Genotyping and Phyloptyping | Phenotype Assays | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERIC-PCR | RAPD-PCR Profile | XbaI-PFGE Profile | Phylo-Group | β-glu | Conju-Gation | Biofilm | Curli | Cellulose | ||||
| Profile | Relative Abundance | Log (cfu/g) | LBWS | M9 | ||||||||
| 01.01 | E01 | 100% | 7.56 | R01 | P01 | B1 | + | − | − | − | − | |
| 02.03 | E02 | 8% | 5.22 | R02 | P02 | B2 | + | − | − | − | − | |
| 02.16 | E03 | 92% | 6.28 | R03 | P03 | F | + | + | − | + | + | |
| 03.25 | E04 | 99% | 7.52 | R04 | P04 | B2 | + | + | − | − | − | |
| 03.73 | E05 | 1% | 5.53 | R03 | P05 | A | + | + | + | − | − | |
| 04.05 | E06 | 5% | 4.22 | R05 | P06 | A | + | + | − | + | − | |
| 04.06 | E07 | 70% | 5.37 | R06 | P07 | B2 | + | − | − | − | − | |
| 04.10 | E08 | 25% | 4.92 | R07 | P08 | E | + | − | − | + | + | |
| 05.20 | E07 | 96% | 7.78 | R02 | P09 | B2 | + | − | − | − | + | |
| 05.47 | E09 | 4% | 6.40 | R08 | P10 | B1 | − | + | − | − | − | − |
| 06.16 | E07 | 100% | 7.28 | R07 | P11 | B2 | + | − | − | − | − | |
| 07.09 | E10 | 100% | 6.45 | R04 | P12 | D | + | + | − | + | − | |
| 08.01 | E11 | 18% | 4.91 | R09 | P12 | D | + | + | − | + | − | |
| 08.02 | E12 | 5% | 4.37 | R10 | P13 | A | + | − | + | − | − | |
| 08.10 | E13 | 5% | 4.37 | R11 | P14 | A | + | + | + | + | − | |
| 08.27 | E14 | 36% | 5.20 | R11 | P15 | B1 | − | + | − | + | − | − |
| 08.57 | E15 | 34% | 5.18 | R12 | P16 | F | − | + | − | − | − | − |
| 08.109 | E16 | 2% | 3.86 | R13 | P17 | F | − | + | − | − | − | − |
| 09.13 | E01 | 92% | 7.09 | R14 | P18 | C | + | + | − | + | + | |
| 09.21 | E17 | 4% | 5.73 | R12 | P19 | F | + | − | − | − | − | |
| 09.42 | E07 | 4% | 5.73 | R12 | P09 | B2 | + | − | − | − | − | |
| 10.17 | E04 | 100% | 7.62 | R12 | P04 | B2 | + | + | − | − | − | |
| 11.02 | E18 | 50% | 7.67 | R15 | P20 | A | + | − | − | − | − | |
| 11.04 | E19 | 9% | 6.91 | R10 | P21 | A | + | − | − | − | − | |
| 11.06 | E07 | 37% | 7.54 | R16 | P22 | B2 | + | − | − | − | − | |
| 11.16 | E20 | 4% | 6.57 | R17 | P23 | B2 | + | − | − | − | − | |
| 12.04 | E13 | 71% | 7.43 | R18 | P24 | B2 | + | − | − | + | − | |
| 12.09 | E11 | 29% | 7.04 | R18 | P25 | D | + | − | − | − | − | |
| 13.01 | E01 | 3% | 4.98 | R19 | P26 | B1 | + | − | − | + | − | |
| 13.03 | E11 | 1% | 4.47 | R11 | P27 | D | + | − | − | + | + | |
| 13.20 | E21 | 97% | 6.46 | R13 | P28 | F | − | − | − | − | − | − |
| 14.03 | E22 | 100% | 6.85 | R13 | P29 | E | + | − | − | + | − | |
| 15.01 | E07 | 9% | 6.45 | R13 | P30 | B2 | + | − | − | − | + | |
| 15.02 | E01 | 87% | 7.45 | R11 | P31 | B1 | + | + | − | + | + | |
| 15.13 | E11 | 4% | 6.15 | R20 | P32 | D | + | − | − | + | − | |
| 16.04 | E21 | 1% | 5.74 | R21 | P33 | F | + | − | − | − | − | |
| 16.15 | E01 | 1% | 5.74 | R22 | P34 | B1 | + | + | − | + | − | |
| 16.27 | E21 | 98% | 7.73 | R23 | P33 | F | − | − | − | − | − | − |
| 17.10 | E23 | 87% | 7.45 | R23 | P35 | A | + | − | − | − | − | |
| 17.18 | E07 | 13% | 6.63 | R24 | P36 | B2 | + | + | − | − | − | |
| 18.03 | E24 | 39% | 6.25 | R25 | P37 | B1 | + | − | + | − | − | |
| 18.09 | E13 | 41% | 6.28 | R25 | P24 | B2 | + | − | − | + | − | |
| 18.18 | E25 | 19% | 5.93 | R13 | P38 | E | + | − | − | − | + | |
| 18.50 | E26 | 1% | 4.70 | R21 | P39 | B1 | − | + | − | − | − | − |
| 19.02 | E07 | 100% | 7.19 | R14 | P40 | B2 | + | + | − | − | − | |
| 20.04 | E27 | 12% | 6.39 | R26 | P41 | B1 | + | − | − | + | − | |
| 20.17 | E28 | 5% | 6.01 | R12 | P42 | C | + | − | − | + | − | |
| 20.27 | E29 | 53% | 7.03 | R27 | P43 | E | + | + | − | + | + | |
| 20.50 | E30 | 22% | 6.65 | R04 | P44 | B1 | − | + | − | − | − | − |
| 20.51 | E31 | 4% | 5.91 | R06 | P44 | B1 | − | + | − | − | − | − |
| 20.71 | E09 | 4% | 5.91 | R06 | P44 | B1 | − | + | − | − | − | − |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raimondi, S.; Righini, L.; Candeliere, F.; Musmeci, E.; Bonvicini, F.; Gentilomi, G.; Starčič Erjavec, M.; Amaretti, A.; Rossi, M. Antibiotic Resistance, Virulence Factors, Phenotyping, and Genotyping of E. coli Isolated from the Feces of Healthy Subjects. Microorganisms 2019, 7, 251. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms7080251
Raimondi S, Righini L, Candeliere F, Musmeci E, Bonvicini F, Gentilomi G, Starčič Erjavec M, Amaretti A, Rossi M. Antibiotic Resistance, Virulence Factors, Phenotyping, and Genotyping of E. coli Isolated from the Feces of Healthy Subjects. Microorganisms. 2019; 7(8):251. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms7080251
Chicago/Turabian StyleRaimondi, Stefano, Lucia Righini, Francesco Candeliere, Eliana Musmeci, Francesca Bonvicini, Giovanna Gentilomi, Marjanca Starčič Erjavec, Alberto Amaretti, and Maddalena Rossi. 2019. "Antibiotic Resistance, Virulence Factors, Phenotyping, and Genotyping of E. coli Isolated from the Feces of Healthy Subjects" Microorganisms 7, no. 8: 251. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms7080251