In Vitro Zika Virus Infection of Human Neural Progenitor Cells: Meta-Analysis of RNA-Seq Assays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Search
2.2. RNA-Seq Data Collection, Processing, and Analysis
2.3. Meta-Analysis
2.4. Gene Ontology Enrichment Analysis
2.5. Reactome Pathway Analysis
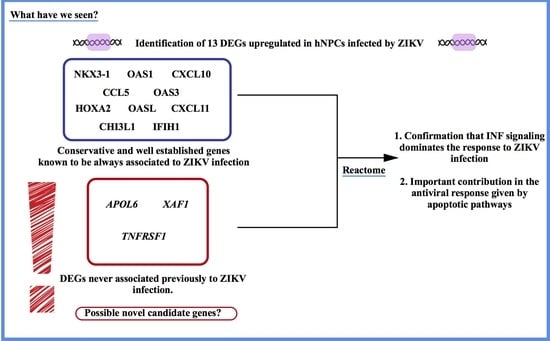
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Azar, S.R.; Weaver, S.C. Vector Competence: What Has Zika Virus Taught Us? Viruses 2019, 11, 867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuler-Faccini, L.; Ribeiro, E.M.; Feitosa, I.M.; Horovitz, D.D.; Cavalcanti, D.P.; Pessoa, A.; Doriqui, M.J.; Neri, J.I.; Neto, J.M.; Wanderley, H.Y.; et al. Possible Association Between Zika Virus Infection and Microcephaly—Brazil, 2015. Morb. Mortal. Wkly. Rep. 2016, 65, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Filho Dde, B.; Martelli, C.M.; Ximenes, R.A.; Araujo, T.V.; Rocha, M.A.; Ramos, R.C.; Dhalia, R.; Franca, R.F.; Marques Junior, E.T.; Rodrigues, L.C. Initial Description of the Presumed Congenital Zika Syndrome. Am. J. Public Health 2016, 106, 598–600. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.A.; Staples, J.E.; Dobyns, W.B.; Pessoa, A.; Ventura, C.V.; Fonseca, E.B.; Ribeiro, E.M.; Ventura, L.O.; Neto, N.N.; Arena, J.F.; et al. Characterizing the Pattern of Anomalies in Congenital Zika Syndrome for Pediatric Clinicians. JAMA Pediatr. 2017, 171, 288–295. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.F.; Choi, G.K.; Yip, C.C.; Cheng, V.C.; Yuen, K.Y. Zika fever and congenital Zika syndrome: An unexpected emerging arboviral disease. J. Infect. 2016, 72, 507–524. [Google Scholar] [CrossRef] [Green Version]
- Ventura, C.V.; Maia, M.; Ventura, B.V.; Linden, V.V.; Araujo, E.B.; Ramos, R.C.; Rocha, M.A.; Carvalho, M.D.; Belfort, R., Jr.; Ventura, L.O. Ophthalmological findings in infants with microcephaly and presumable intra-uterus Zika virus infection. Arq. Bras. Oftalmol. 2016, 79, 1–3. [Google Scholar] [CrossRef]
- De Paula Freitas, B.; de Oliveira Dias, J.R.; Prazeres, J.; Sacramento, G.A.; Ko, A.I.; Maia, M.; Belfort, R., Jr. Ocular Findings in Infants With Microcephaly Associated With Presumed Zika Virus Congenital Infection in Salvador, Brazil. JAMA Ophthalmol. 2016, 134, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Ventura, C.V.; Maia, M.; Bravo-Filho, V.; Gois, A.L.; Belfort, R., Jr. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 2016, 387, 228. [Google Scholar] [CrossRef] [Green Version]
- O’Rahilly, R.; Muller, F. Developmental stages in human embryos: Revised and new measurements. Cells Tissues Org. 2010, 192, 73–84. [Google Scholar] [CrossRef]
- Martinez-Cerdeno, V.; Noctor, S.C. Neural Progenitor Cell Terminology. Front. Neuroanat. 2018, 12, 104. [Google Scholar] [CrossRef]
- Hammack, C.; Ogden, S.C.; Madden, J.C., Jr.; Medina, A.; Xu, C.; Phillips, E.; Son, Y.; Cone, A.; Giovinazzi, S.; Didier, R.A.; et al. Zika Virus Infection Induces DNA Damage Response in Human Neural Progenitors That Enhances Viral Replication. J. Virol. 2019, 93, e00638-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddique, R.; Liu, Y.; Nabi, G.; Sajjad, W.; Xue, M.; Khan, S. Zika Virus Potentiates the Development of Neurological Defects and Microcephaly: Challenges and Control Strategies. Front. Neurol. 2019, 10, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa-Fernandes, L.; Cugola, F.R.; Russo, F.B.; Kawahara, R.; de Melo Freire, C.C.; Leite, P.E.C.; Bassi Stern, A.C.; Angeli, C.B.; de Oliveira, D.B.L.; Melo, S.R.; et al. Zika Virus Impairs Neurogenesis and Synaptogenesis Pathways in Human Neural Stem Cells and Neurons. Front. Cell Neurosci. 2019, 13, 64. [Google Scholar] [CrossRef]
- Lima, M.C.; de Mendonca, L.R.; Rezende, A.M.; Carrera, R.M.; Anibal-Silva, C.E.; Demers, M.; D’Aiuto, L.; Wood, J.; Chowdari, K.V.; Griffiths, M.; et al. The Transcriptional and Protein Profile From Human Infected Neuroprogenitor Cells Is Strongly Correlated to Zika Virus Microcephaly Cytokines Phenotype Evidencing a Persistent Inflammation in the CNS. Front. Immunol. 2019, 10, 1928. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Costa-Silva, J.; Domingues, D.; Lopes, F.M. RNA-Seq differential expression analysis: An extended review and a software tool. PLoS ONE 2017, 12, e0190152. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Kumar, V.; Olson, A.; Ware, D. Reviving the Transcriptome Studies: An Insight Into the Emergence of Single-Molecule Transcriptome Sequencing. Front. Genet. 2019, 10, 384. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Stephens, R.M.; Meltzer, P.S.; Davis, S.R. SRAdb: Query and use public next-generation sequencing data from within R. BMC Bioinformat. 2013, 14, 19. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.r-project.org/ (accessed on 3 July 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Pagès, H.; Obenchain, V.; Hayden, N. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R Package Vers. 2016, 1, 677–689. [Google Scholar]
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Hong, F.; Breitling, R.; McEntee, C.W.; Wittner, B.S.; Nemhauser, J.L.; Chory, J. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 2006, 22, 2825–2827. [Google Scholar] [CrossRef] [Green Version]
- Del Carratore, F.; Jankevics, A.; Eisinga, R.; Heskes, T.; Hong, F.; Breitling, R. RankProd 2.0: A refactored bioconductor package for detecting differentially expressed features in molecular profiling datasets. Bioinformatics 2017, 33, 2774–2775. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Park, Y.K.; Yoon, C.H.; Kim, K.; Kim, K.C. Meta-analysis of gene expression profiles in long-term non-progressors infected with HIV-1. BMC Med. Genomics 2019, 12, 3. [Google Scholar] [CrossRef]
- Rue-Albrecht, K. GOexpress: Visualise Microarray and RNAseq Data Using Gene Ontology Annotations. R Package Version 1.18.0. Available online: https://github.com/kevinrue/GOexpress (accessed on 7 January 2020).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Hammack, C.; Ogden, S.C.; Cheng, Y.; Lee, E.M.; Wen, Z.; Qian, X.; Nguyen, H.N.; Li, Y.; Yao, B.; et al. Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res. 2016, 44, 8610–8620. [Google Scholar] [CrossRef]
- McGrath, E.L.; Rossi, S.L.; Gao, J.; Widen, S.G.; Grant, A.C.; Dunn, T.J.; Azar, S.R.; Roundy, C.M.; Xiong, Y.; Prusak, D.J.; et al. Differential Responses of Human Fetal Brain Neural Stem Cells to Zika Virus Infection. Stem Cell Rep. 2017, 8, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Caires-Junior, L.C.; Goulart, E.; Melo, U.S.; Araujo, B.H.S.; Alvizi, L.; Soares-Schanoski, A.; de Oliveira, D.F.; Kobayashi, G.S.; Griesi-Oliveira, K.; Musso, C.M.; et al. Discordant congenital Zika syndrome twins show differential in vitro viral susceptibility of neural progenitor cells. Nat. Commun. 2018, 9, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, M.M.; Abate-Shen, C. Roles of the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Dev. Dyn. 2003, 228, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef] [Green Version]
- Coffman, F.D. Chitinase 3-Like-1 (CHI3L1): A putative disease marker at the interface of proteomics and glycomics. Crit. Rev. Clin. Lab. Sci. 2008, 45, 531–562. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, H.; Gad, H.H.; Eskildsen-Larsen, S.; Despres, P.; Hartmann, R. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 2011, 31, 41–47. [Google Scholar] [CrossRef]
- Whelan, J.N.; Li, Y.; Silverman, R.H.; Weiss, S.R. Zika Virus Production Is Resistant to RNase L Antiviral Activity. J. Virol. 2019, 93, e00313-19. [Google Scholar] [CrossRef] [Green Version]
- Urcuqui-Inchima, S.; Cabrera, J.; Haenni, A.L. Interplay between dengue virus and Toll-like receptors, RIG-I/MDA5 and microRNAs: Implications for pathogenesis. Antivir. Res. 2017, 147, 47–57. [Google Scholar] [CrossRef]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Luplertlop, N.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of Zika Virus Infection in Human Skin Cells. J. Virol. 2015, 89, 8880–8896. [Google Scholar] [CrossRef] [Green Version]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lu, H.; Jiang, Z.; Pastuszyn, A.; Hu, C.A. Apolipoprotein l6, a novel proapoptotic Bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol. Cancer Res. 2005, 3, 21–31. [Google Scholar] [PubMed]
- Plenchette, S.; Cheung, H.H.; Fong, W.G.; LaCasse, E.C.; Korneluk, R.G. The role of XAF1 in cancer. Curr. Opin. Investig. Drugs 2007, 8, 469–476. [Google Scholar] [PubMed]
- Siebert, S.; Fielding, C.A.; Williams, B.D.; Brennan, P. Mutation of the extracellular domain of tumor necrosis factor receptor 1 causes reduce NF-kappaB activation due to decreased surface expression. FEBS Lett. 2005, 579, 5193–5198. [Google Scholar] [CrossRef] [Green Version]
- Shui, J.W.; Kronenberg, M. HVEM is a TNF Receptor with Multiple Regulatory Roles in the Mucosal Immune System. Immune Netw. 2014, 14, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Commins, S.P.; Borish, L.; Steinke, J.W. Immunologic messenger molecules: Cytokines, interferons, and chemokines. J. Allergy Clin. Immunol. 2010, 125, S53–S72. [Google Scholar] [CrossRef]
- Morimoto, K.; Nakajima, K. Role of the Immune System in the Development of the Central Nervous System. Front. Neurosci. 2019, 13, 916. [Google Scholar] [CrossRef] [Green Version]

| SRA | Title | Samples | Replicates | Main Results |
|---|---|---|---|---|
| SRP073493 | Molecular Signatures Associated with ZIKV Exposure in Human Cortical Neural Progenitors [32] | Three infected (two with African and one with Asian lineage); two non-infected | Two per sample | The RNA-Seq extraction was gone in 56 hpi for African lineage and 64 hpi for Asian lineage. MOI of 0.2 and 0.4. DEGs include TP53. |
| SRP096367 | Differential Responses of Human Fetal Brain Neural Stem Cells to Zika Virus Infection [33] | Three infected with Asian or African lineage; three non-infected | Three per sample | Usage of isolates from Mexico (Asian lineage), Cambodia (Asian lineage), and Senegal strains (African lineage). Following 120 hpi to RNA-Seq extraction. MOI of 0.1 and 1. The DEGs found were FAS, SOX1, and TUBB3. |
| SRP114529 | RNA-seq of hiPSCs-Derived NPCs from Three Pairs of Dizygotic Discordant Twins for Congenital Zika Syndrome [34] | Three infected with Asian lineage; three non-infected | One per sample | Brazilian strain (Asian lineage) used at a MOI of 0.01 and 0.1. RNA-Seq extracted 96 hpi. Indentified DEGs included DEPDC5, GPR108, MICAL3, OR12D2, OR4K5, PHF2, SLC6A18, and TTC16. |
| Gene | Rank Product | Fold Change | log2 (Fold Change) | p-Value | pfp |
|---|---|---|---|---|---|
| (Control/ZIKV-Infected) | |||||
| OAS1 | 334.5 | 0.228 | −2.1329 | 1.874 × 10−9 | 0.0001 |
| CXCL10 | 350.9 | 0.3021 | −1.7269 | 2.664 × 10−9 | 0.00004 |
| OASL | 840.8 | 0.3739 | −1.4193 | 0.000001 | 0.0088 |
| CCL5 | 880.8 | 0.2667 | −1.9067 | 0.000002 | 0.0095 |
| CXCL11 | 997.5 | 0.1634 | −2.6135 | 0.000004 | 0.0153 |
| TNFRSF1 | 1001 | 0.2218 | −2.1727 | 0.000004 | 0.0137 |
| IFIH1 | 1085 | 0.2048 | −2.2877 | 0.000006 | 0.0205 |
| OAS3 | 1150 | 0.3442 | −1.5387 | 0.000009 | 0.0266 |
| CHI3L1 | 1158 | 0.3843 | −1.3797 | 0.00001 | 0.0254 |
| NKX3-1 | 1164 | 0.3466 | −1.5287 | 0.00001 | 0.0241 |
| APOL6 | 1203 | 0.1532 | −2.7065 | 0.00001 | 0.0273 |
| XAF1 | 1251 | 0.283 | −1.8213 | 0.00002 | 0.0323 |
| HOXA2 | 1314 | 0.4441 | −1.1710 | 0.00002 | 0.0383 |
| Rank | GO Term | Genes in GO Term | Genes of GO Term Present in Data | Adjusted p-Value | GO Term Description |
|---|---|---|---|---|---|
| 23 | GO:0043066 | 541 | 526 | 0.00071 | Negative regulation of apoptotic process |
| 49 | GO:0009615 | 109 | 109 | 0.00570 | Response to virus |
| 57 | GO:0006915 | 693 | 666 | 0.00617 | Apoptotic process |
| 64 | GO:0016032 | 476 | 460 | 0.00662 | Viral process |
| 69 | GO:0033209 | 118 | 117 | 0.00749 | Tumor necrosis factor-mediated signaling pathway |
| 125 | GO:1902237 | 11 | 11 | 0.01385 | Positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway |
| 132 | GO:1901216 | 41 | 41 | 0.01523 | Positive regulation of neuron death |
| 142 | GO:0050727 | 81 | 81 | 0.01576 | Regulation of inflammatory response |
| 152 | GO:0006954 | 386 | 372 | 0.01612 | Inflammatory response |
| 257 | GO:0006959 | 56 | 56 | 0.02419 | Humoral immune response |
| 282 | GO:0043065 | 375 | 360 | 0.02500 | Positive regulation of apoptotic process |
| 328 | GO:0051607 | 201 | 195 | 0.02596 | Defense response to virus |
| 546 | GO:0042771 | 29 | 29 | 0.03671 | Intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator |
| 570 | GO:0034612 | 32 | 32 | 0.03800 | Response to tumor necrosis factor |
| 576 | GO:0002523 | 9 | 9 | 0.03854 | Leukocyte migration involved in inflammatory response |
| 639 | GO:0097194 | 18 | 18 | 0.04256 | Execution phase of apoptosis |
| 641 | GO:0045089 | 25 | 25 | 0.04259 | Positive regulation of innate immune response |
| 683 | GO:0002741 | 8 | 8 | 0.04391 | Positive regulation of cytokine secretion involved in immune response |
| 704 | GO:0002437 | 15 | 15 | 0.04585 | Inflammatory response to antigenic stimulus |
| 805 | GO:0002827 | 9 | 9 | 0.04832 | Positive regulation of T-helper 1 type immune response |
| Reactome ID | Description | p-Value | Adjusted p-Value | Gene Symbols |
|---|---|---|---|---|
| R-HSA-1169410 | Antiviral mechanism by IFN-stimulated genes | 6.50 × 10−5 | 0.0005 | OAS1/OASL/OAS3 |
| R-HSA-373076 | Class A/1 (Rhodopsin-like receptors) | 0.0039 | 0.0135 | CXCL10/CCL5/CXCL11 |
| R-HSA-375276 | Peptide ligand-binding receptors | 0.0008 | 0.0039 | CXCL10/CCL5/CXCL11 |
| R-HSA-380108 | Chemokine receptors bind chemokines | 1.39 × 10−5 | 0.0002 | CXCL10/CCL5/CXCL11 |
| R-HSA-418594 | G alpha (i) signaling events | 0.0073 | 0.0228 | CXCL10/CCL5/CXCL11 |
| R-HSA-500792 | GPCR ligand binding | 0.0101 | 0.0282 | CXCL10/CCL5/CXCL11 |
| R-HSA-6783783 | Interleukin-10 signaling | 0.0010 | 0.0041 | CXCL10/CCL5 |
| R-HSA-877300 | Interferon gamma signaling | 9.87 × 10−5 | 0.0005 | OAS1/OASL/OAS3 |
| R-HSA-909733 | Interferon alpha/beta signaling | 5.21 × 10−7 | 1.46 × 10−5 | OAS1/OASL/OAS3/XAF1 |
| R-HSA-913531 | Interferon Signaling | 3.56 × 10−5 | 0.0003 | OAS1/OASL/OAS3/XAF1 |
| R-HSA-918233 | TRAF3-dependent IRF activation pathway | 0.0144 | 0.0336 | IFIH1 |
| R-HSA-933543 | NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 | 0.0127 | 0.0315 | IFIH1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gratton, R.; Tricarico, P.M.; Agrelli, A.; Colaço da Silva, H.V.; Coêlho Bernardo, L.; Crovella, S.; Campos Coelho, A.V.; Rodrigues de Moura, R.; Cavalcanti Brandão, L.A. In Vitro Zika Virus Infection of Human Neural Progenitor Cells: Meta-Analysis of RNA-Seq Assays. Microorganisms 2020, 8, 270. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8020270
Gratton R, Tricarico PM, Agrelli A, Colaço da Silva HV, Coêlho Bernardo L, Crovella S, Campos Coelho AV, Rodrigues de Moura R, Cavalcanti Brandão LA. In Vitro Zika Virus Infection of Human Neural Progenitor Cells: Meta-Analysis of RNA-Seq Assays. Microorganisms. 2020; 8(2):270. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8020270
Chicago/Turabian StyleGratton, Rossella, Paola Maura Tricarico, Almerinda Agrelli, Heverton Valentim Colaço da Silva, Lucas Coêlho Bernardo, Sergio Crovella, Antonio Victor Campos Coelho, Ronald Rodrigues de Moura, and Lucas André Cavalcanti Brandão. 2020. "In Vitro Zika Virus Infection of Human Neural Progenitor Cells: Meta-Analysis of RNA-Seq Assays" Microorganisms 8, no. 2: 270. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8020270