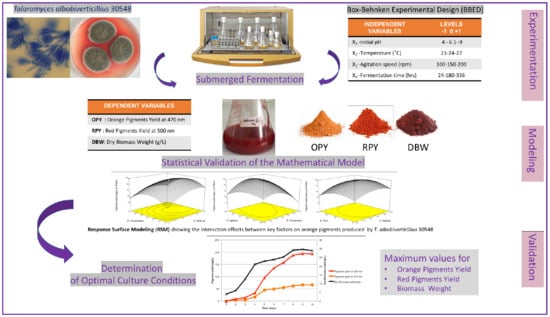
Statistical Optimization of the Physico-Chemical Parameters for Pigment Production in Submerged Fermentation of Talaromyces albobiverticillius 30548
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism and Maintenance
2.2. Preparation of Pre-Culture
2.3. Fermentation Conditions
2.4. Spectrophotometric Quantification of Extracellular Pigments
2.5. Dry Biomass Concentration
2.6. Experimental Design and Statistical Analysis (BBED and RSM)
2.7. Optimization and Validation
3. Results
3.1. Box–Behnken Experimental Design Analysis
3.2. Development of Second-Order Polynomial Models
3.3. Determination of Second-Order Polynomial Equations
3.4. Statistical Analysis
3.5. Effect of Process Variables on Pigment Yield
3.5.1. Combined Effect of pH, Temperature, Agitation Speed, and Time on Orange Pigment Yield (OPY)
3.5.2. Combined Effects of pH, Temperature, Agitation Speed, and Time on Red Pigment Yield (RPY)
3.5.3. Combined Effects of pH, Temperature, Agitation Speed, and Time on Dry Biomass Weight (DBW)
3.6. Multi Response Optimization
3.7. Validation of the Optimized Condition
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A


Appendix B
| Source | DF | Orange Pigment Yield | Red Pigment Yield | Dry Biomass Weight | |||
|---|---|---|---|---|---|---|---|
| RC | p Value * | RC | p Value * | RC | p Value * | ||
| Model | 14 | 1.74 | <0.0001 | 1.92 | <0.0001 | 6.80 | <0.0001 |
| X1 | 1 | 0.35 | <0.0001 | 0.28 | <0.0001 | 0.77 | <0.0001 |
| X2 | 1 | −0.11 | 0.0009 | −0.090 | 0.0058 | −0.77 | <0.0001 |
| X3 | 1 | 0.27 | <0.0001 | 0.20 | <0.0001 | 1.24 | <0.0001 |
| X4 | 1 | −0.029 | 0.2767 | −0.015 | 0.5901 | −0.17 | 0.1837 |
| X12 | 1 | −0.11 | 0.0245 | −0.087 | 0.0927 | −0.24 | 0.2788 |
| X13 | 1 | −0.19 | 0.0009 | −0.17 | 0.0031 | −0.02 | 0.9224 |
| X14 | 1 | 0.11 | 0.0245 | 0.075 | 0.1394 | −0.61 | 0.0117 |
| X23 | 1 | 0.41 | <0.0001 | 0.31 | <0.0001 | 1.18 | <0.0001 |
| X24 | 1 | 0.000 | 1.000 | −0.019 | 0.7011 | 0.55 | 0.0210 |
| X34 | 1 | 0.10 | 0.0418 | 0.071 | 0.1615 | 0.17 | 0.4359 |
| X12 | 1 | −0.42 | <0.0001 | −0.33 | <0.0001 | −1.02 | <0.0001 |
| X22 | 1 | −0.68 | <0.0001 | −0.54 | <0.0001 | −2.31 | <0.0001 |
| X32 | 1 | −0.35 | <0.0001 | −0.31 | <0.0001 | −1.35 | <0.0001 |
| X42 | 1 | −0.38 | <0.0001 | −0.32 | <0.0001 | −0.97 | <0.0001 |
| R2 | 0.9853 | 0.9734 | 0.9714 | ||||
| AdjR2 | 0.9706 | 0.9468 | 0.9429 | ||||
| PreR2 | 0.9172 | 0.8469 | 0.8445 | ||||
| CV% | 9.12 | 7.41 | 9.41 | ||||
| Adeq. Pre. | 28.30 | 21.04 | 22.71 | ||||
References
- Weiss, B. Synthetic Food Colors and Neurobehavioral Hazards: The View from Environmental Health Research. Environ. Health Perspect. 2012, 120, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Dufossé, L. Microbial production of food grade pigments. Food Technol. Biotechnol. 2006, 44, 313–323. [Google Scholar]
- Dufosse, L. Anthraquinones, the Dr Jekyll and Mr Hyde of the food pigment family. Food Res. Int. 2014, 65, 132–136. [Google Scholar] [CrossRef]
- Kumar, A.; Vishwakarma, H.S.; Singh, J.; Dwivedi, S.; Kumar, M. Microbial pigments: Production and their applications in various industries. IJPCBS 2015, 5, 203–212. [Google Scholar]
- Kamarudin, K.R. Microbial production of food grade pigments–screening and metabolic pathway analysis. Food Technol. Biotechnol. 2006, 44, 13–23. [Google Scholar]
- Caro, Y.; Anamale, L.; Fouillaud, M.; Laurent, P.; Petit, T.; Dufosse, L. Natural hydroxyanthraquinoid pigments as potent food grade colorants: An overview. Nat. Prod. Bioprospect. 2012, 2, 174–193. [Google Scholar] [CrossRef]
- Duran, N.; Teixeira, M.F.; De Conti, R.; Esposito, E. Ecological-friendly pigments from fungi. Crit. Rev. Food Sci. Nutr. 2002, 42, 53–66. [Google Scholar] [CrossRef]
- Ventura, S.P.; Santos-Ebinuma, V.C.; Pereira, J.F.; Teixeira, M.F.; Pessoa, A.; Coutinho, J.A. Isolation of natural red colorants from fermented broth using ionic liquid-based aqueous two-phase systems. J. Ind. Microbiol. Biotechnol. 2013, 40, 507–516. [Google Scholar] [CrossRef]
- Pastre, R.; Marinho, A.M.; Rodrigues-Filho, E.; Souza, A.Q.; Pereira, J.O. Diversity of polyketides produced by Penicillium species isolated from Melia azedarach and Murraya paniculata. Química Nova 2007, 30, 1867–1871. [Google Scholar] [CrossRef] [Green Version]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Osmanova, N.; Schultze, W.; Ayoub, N. Azaphilones: A class of fungal metabolites with diverse biological activities. Phytochem. Rev. 2010, 9, 315–342. [Google Scholar] [CrossRef]
- Kongruang, S. Growth kinetics of biopigment production by Thai isolated Monascus purpureus in a stirred tank bioreactor. J. Ind. Microbiol. Biotechnol. 2011, 38, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.F.; Martins, M.S.; Da Silva, J.; Kirsch, L.S.; Fernandes, O.C.; Carneiro, A.L.; De Conti, R.; Durán, N. Amazonian biodiversity: Pigments from Aspergillus and Penicillium–characterizations, antibacterial activities and their toxicities. Curr. Trends Biotechnol. Pharm. 2012, 6, 300–311. [Google Scholar]
- Torres, F.A.E.; Zaccarim, B.R.; De Lencastre Novaes, L.C.; Jozala, A.F.; Santos, C.A.d.; Teixeira, M.F.S.; Santos-Ebinuma, V.C. Natural colorants from filamentous fungi. Appl. Microbiol. Biotechnol. 2016, 100, 2511–2521. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, I.; Kim, S.-K. Immense Essence of Excellence: Marine Microbial Bioactive Compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [Green Version]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef]
- Imhoff, J.F. Natural products from marine fungi—Still an underrepresented resource. Mar. drugs 2016, 14, 19. [Google Scholar] [CrossRef]
- Yilmaz, N.; Houbraken, J.; Hoekstra, E.; Frisvad, J.C.; Visagie, C.; Samson, R. Delimitation and characterisation of Talaromyces purpurogenus and related species. Pers. Mol. Phylogeny Evol. Fungi 2012, 29, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Frisvad, J.C.; Yilmaz, N.; Thrane, U.; Rasmussen, K.B.; Houbraken, J.; Samson, R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 2013, 8, e84102. [Google Scholar] [CrossRef] [Green Version]
- Mapari, S.A.; Meyer, A.S.; Thrane, U.; Frisvad, J.C. Identification of potentially safe promising fungal cell factories for the production of polyketide natural food colorants using chemotaxonomic rationale. Microbial Cell Factories 2009, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Nicoletti, R.; Trincone, A. Bioactive compounds produced by strains of Penicillium and Talaromyces of marine origin. Mar. Drugs 2016, 14, 37. [Google Scholar] [CrossRef] [Green Version]
- Caro, Y.; Venkatachalam, M.; Lebeau, J.; Fouillaud, M.; Dufossé, L. Pigments and colorants from filamentous fungi. In Fungal Metabolites, 1st ed.; Merillon, J.-M., Ramawat, K.G., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–70. [Google Scholar]
- Musoni, M.; Destain, J.; Thonart, P.; Bahama, J.-B.; Delvigne, F. Bioreactor design and implementation strategies for the cultivation of filamentous fungi and the production of fungal metabolites: From traditional methods to engineered systems/Conception de bioréacteurs et mise en oeuvre de stratégies pour la culture de champignons filamenteux et la production de métabolites d’origine fongique: Des méthodes traditionnelles aux technologies actuelles. Biotechnol. Agron. Société Environ. 2015, 19, 430. [Google Scholar]
- General, T.; Kim, H.-J.; Prasad, B.; Ngo, H.T.A.; Vadakedath, N.; Cho, M.-G. Fungal utilization of a known and safe macroalga for pigment production using solid-state fermentation. J. Appl. Phycol. 2014, 26, 1547–1555. [Google Scholar] [CrossRef]
- Hajjaj, H.; Blanc, P.; Goma, G.; Francois, J. Sampling techniques and comparative extraction procedures for quantitative determination of intra-and extracellular metabolites in filamentous fungi. FEMS Microbiol. Lett. 1998, 164, 195–200. [Google Scholar] [CrossRef]
- Santos-Ebinuma, V.C.; Roberto, I.C.; Teixeira, M.F.S.; Pessoa, A., Jr. Improvement of submerged culture conditions to produce colorants by Penicillium purpurogenum. Braz. J. Microbiol. 2014, 45, 731–742. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.-K.; Park, N.-H.; Piao, H.Y.; Chung, W.-J. Production of red pigments by Monascus purpureus in submerged culture. Biotechnol. Bioprocess Eng. 2001, 6, 341–346. [Google Scholar] [CrossRef]
- Méndez, A.; Pérez, C.; Montañéz, J.C.; Martínez, G.; Aguilar, C.N. Red pigment production by Penicillium purpurogenum GH2 is influenced by pH and temperature. J. Zhejiang Univ. Sci. B 2011, 12, 961–968. [Google Scholar] [CrossRef] [Green Version]
- Orozco, S.F.B.; Kilikian, B.V. Effect of pH on citrinin and red pigments production by Monascus purpureus CCT3802. World J. Microbiol. Biotechnol. 2008, 24, 263–268. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, J.; Huang, Y.; Xin, Q.; Wang, Z. Diversifying of chemical structure of native Monascus pigments. Front. Microbiol. 2018, 9, 3143. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Studies Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; He, L.; Hu, M. Optimized ultrasonic-assisted extraction of flavonoids from Prunella vulgaris L. and evaluation of antioxidant activities in vitro. Innov. Food Sci. Emerg. Technol. 2011, 12, 18–25. [Google Scholar] [CrossRef]
- Yang, Z.; Zhai, W. Optimization of microwave-assisted extraction of anthocyanins from purple corn (Zea mays L.) cob and identification with HPLC–MS. Innov. Food Sci. Emerg. Technol. 2010, 11, 470–476. [Google Scholar] [CrossRef]
- Parajó, J.; Santos, V.; Domínguez, H.; Vázquez, M. NH4OH-Based pretreatment for improving the nutritional quality of single-cell protein (SCP). Appl. Biochem. Biotechnol. 1995, 55, 133–149. [Google Scholar] [CrossRef]
- Huang, W.; Xue, A.; Niu, H.; Jia, Z.; Wang, J. Optimised ultrasonic-assisted extraction of flavonoids from Folium eucommiae and evaluation of antioxidant activity in multi-test systems in vitro. Food Chem. 2009, 114, 1147–1154. [Google Scholar] [CrossRef]
- Hayouni, E.A.; Abedrabba, M.; Bouix, M.; Hamdi, M. The effects of solvents and extraction method on the phenolic contents and biological activities in vitro of Tunisian Quercus coccifera L. and Juniperus phoenicea L. fruit extracts. Food Chem. 2007, 105, 1126–1134. [Google Scholar] [CrossRef]
- Ge, Y.; Ni, Y.; Yan, H.; Chen, Y.; Cai, T. Optimization of the supercritical fluid extraction of natural vitamin E from wheat germ using response surface methodology. J. Food Sci. 2002, 67, 239–243. [Google Scholar] [CrossRef]
- Sehrawat, R.; Panesar, P.S.; Swer, T.L.; Kumar, A. Response surface methodology (RSM) mediated interaction of media concentration and process parameters for the pigment production by Monascus purpureus MTCC 369 under solid state fermentation. Pigment. Resin Technol. 2017, 46, 14–20. [Google Scholar] [CrossRef]
- Venkatachalam, M.; Magalon, H.; Dufossé, L.; Fouillaud, M. Production of pigments from the tropical marine-derived fungi Talaromyces albobiverticillius: New resources for natural red-colored metabolites. J. Food Compos. Anal. 2018, 70, 35–48. [Google Scholar] [CrossRef]
- Bas, D.; Boyaci, I.H. Modeling and optimization I: Usability of response surface methodology. J. Food Eng. 2007, 78, 836–845. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef]
- Ferreira, S.C.; Bruns, R.; Ferreira, H.; Matos, G.; David, J.; Brandao, G.; Da Silva, E.P.; Portugal, L.; Dos Reis, P.; Souza, A. Box-Behnken design: An alternative for the optimization of analytical methods. Anal. Chim. Acta 2007, 597, 179–186. [Google Scholar] [CrossRef]
- Karichappan, T.; Venkatachalam, S.; Jeganathan, P.M. Analysis of efficiency of Bacillus subtilis to treat bagasse based paper and pulp industry wastewater-A novel approach. J. Korean Chem. Soc. 2014, 58, 198–204. [Google Scholar] [CrossRef]
- Derringer, G.; Suich, R. Simultaneous Optimization of Several Response Variables. J. Qual. Technol. 1980, 12, 214–219. [Google Scholar] [CrossRef]
- Ekren, O.; Ekren, B.Y. Size optimization of a PV/wind hybrid energy conversion system with battery storage using response surface methodology. Appl. Energy 2008, 85, 1086–1101. [Google Scholar] [CrossRef]
- Venkatachalam, M.; Gérard, L.; Milhau, C.; Vinale, F.; Dufossé, L.; Fouillaud, M. Salinity and Temperature Influence Growth and Pigment Production in the Marine-Derived Fungal Strain Talaromyces albobiverticillius 30548. Microorganisms 2019, 7, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afshari, M.; Shahidi, F.; Mortazavi, S.A.; Tabatabai, F.; Es’ haghi, Z. Investigating the influence of pH, temperature and agitation speed on yellow pigment production by Penicillium aculeatum ATCC 10409. Nat. Prod. Res. 2015, 29, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Park, J.; Hwang, H.; Kim, S.; Choi, J.; Yun, J. Production of red pigment by submerged culture of Paecilomyces sinclairii. Lett. Appl. Microbiol. 2002, 35, 195–202. [Google Scholar] [CrossRef]
- Bae, J.-T.; Sinha, J.; Park, J.-P.; Song, C.-H.; Yun, J.-W. Optimization of submerged culture conditions for exo-biopolymer production by Paecilomyces japonica. J. Microbiol. Biotechnol. 2000, 10, 482–487. [Google Scholar]
- Babitha, S. Microbial Pigments. In Biotechnology for Agro-Industrial Residues Utilisation: Utilisation of Agro-Residues; Singh nee’ Nigam, P., Pandey, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 147–162. [Google Scholar]
- Chadni, Z.; Rahaman, M.H.; Jerin, I.; Hoque, K.; Reza, M.A. Extraction and optimisation of red pigment production as secondary metabolites from Talaromyces verruculosus and its potential use in textile industries. Mycology 2017, 8, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.; Lin, Y.; Koehler, P. Regulation of growth and pigmentation of Monascus purpureus by carbon and nitrogen concentrations. Mycologia 1981, 649–654. [Google Scholar] [CrossRef]
- Yongsmith, B.; Tabloka, W.; Yongmanitchai, W.; Bavavoda, R. Culture conditions for yellow pigment formation by Monascus sp. KB 10 grown on cassava medium. World J. Microbiol. Biotechnol. 1993, 9, 85–90. [Google Scholar] [CrossRef]
- Babitha, S.; Soccol, C.R.; Pandey, A. Solid-state fermentation for the production of Monascus pigments from jackfruit seed. Bioresour. Technol. 2007, 98, 1554–1560. [Google Scholar] [CrossRef]
- Gunasekaran, S.; Poorniammal, R. Optimization of fermentation conditions for red pigment production from Penicillium sp. under submerged cultivation. Afr. J. Biotechnol. 2008, 7, 1894–1898. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Johns, M.R. Effect of pH and nitrogen source on pigment production by Monascus purpureus. Appl. Microbiol. Biotechnol. 1993, 40, 132–138. [Google Scholar] [CrossRef]
- Jafari, A.; Sarrafzadeh, M.; Alemzadeh, I.; Vosoughi, M. Effect of stirrer speed and aeration rate on the production of glucose oxidase by Aspergillus niger. J. Biol. Sci. 2007, 7, 270–275. [Google Scholar]
- Mohamed, M.S.; Mohamad, R.; Manan, M.A.; Ariff, A.B. Enhancement of red pigment production by Monascus purpureus FTC 5391 through retrofitting of helical ribbon impeller in stirred-tank fermenter. Food Bioprocess. Technol. 2012, 5, 80–91. [Google Scholar] [CrossRef]
- Venkatachalam, M.; Zelena, M.; Cacciola, F.; Ceslova, L.; Girard-Valenciennes, E.; Clerc, P.; Dugo, P.; Mondello, L.; Fouillaud, M.; Rotondo, A. Partial characterization of the pigments produced by the marine-derived fungus Talaromyces albobiverticillius 30548. Towards a new fungal red colorant for the food industry. J. Food Compos. Anal. 2018, 67, 38–47. [Google Scholar] [CrossRef]




| Variables | Symbol | Coded and Actual Values | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| pH | X1 | 4 | 6.5 | 9 |
| Temperature (°C) | X2 | 21 | 24 | 27 |
| Agitation speed (rpm) | X3 | 100 | 150 | 200 |
| Fermentation time (h) | X4 | 24 | 168 | 336 |
| Exp. Run | pH | Temperature (°C) | Agitation Speed (rpm) | Fermentation Time (h) | Orange Pigment Yield (g/L1) | Red Pigment Yield (g/L2) | Dry Biomass Weight (g/L) |
|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0.30 | 0.83 | 3.35 |
| 2 | 0 | 0 | 1 | −1 | 1.15 | 1.44 | 5.38 |
| 3 | 0 | 1 | 0 | 1 | 0.30 | 0.82 | 1.97 |
| 4 | −1 | 0 | 0 | 1 | 0.70 | 1.08 | 3.05 |
| 5 | 0 | −1 | 0 | −1 | 0.90 | 1.23 | 3.76 |
| 6 | 0 | 1 | 1 | 0 | 1.10 | 1.37 | 6.17 |
| 7 | −1 | 0 | 0 | −1 | 0.65 | 1.04 | 2.39 |
| 8 | 0 | 0 | 0 | 0 | 1.25 | 1.47 | 5.47 |
| 9 | 0 | 0 | 0 | 0 | 0.65 | 0.95 | 3.34 |
| 10 | 0 | 1 | −1 | 0 | 1.20 | 1.48 | 6.05 |
| 11 | 1 | 1 | 0 | 0 | 0.40 | 0.85 | 4.83 |
| 12 | 0 | 0 | 0 | 0 | 1.40 | 1.68 | 5.10 |
| 13 | 0 | 0 | 0 | 0 | 1.00 | 1.31 | 3.33 |
| 14 | 0 | −1 | −1 | 0 | −0.10 | 0.43 | 0.21 |
| 15 | −1 | 0 | 1 | 0 | 0.65 | 1.03 | 3.75 |
| 16 | −1 | −1 | 0 | 0 | 1.20 | 1.39 | 5.34 |
| 17 | 0 | 1 | 0 | −1 | 0.10 | 0.57 | 2.66 |
| 18 | 1 | 0 | −1 | 0 | 1.20 | 1.48 | 4.28 |
| 19 | 0 | −1 | 1 | 0 | 1.20 | 1.50 | 4.61 |
| 20 | 1 | 0 | 1 | 0 | 1.55 | 1.73 | 6.15 |
| 21 | 1 | 0 | 0 | −1 | 0.85 | 1.18 | 5.21 |
| 22 | 0 | −1 | 0 | 1 | 0.70 | 1.12 | 2.10 |
| 23 | 0 | 0 | −1 | −1 | 0.75 | 1.12 | 3.87 |
| 24 | 1 | −1 | 0 | 0 | 0.60 | 0.98 | 2.93 |
| 25 | −1 | 1 | 0 | 0 | 1.70 | 1.92 | 6.67 |
| 26 | 1 | 0 | 0 | 1 | 1.71 | 1.92 | 6.7 |
| 27 | −1 | 0 | −1 | 0 | 1.76 | 1.93 | 6.79 |
| 28 | 0 | 0 | 1 | 1 | 1.77 | 1.93 | 7.17 |
| 29 | 0 | 0 | −1 | 1 | 1.75 | 1.93 | 6.65 |
| Source | Std. Dev. | R2 | Adjusted R2 | Predicted R2 | Press | Remarks |
|---|---|---|---|---|---|---|
| Model summary statistics for Orange Pigment Yield | ||||||
| Linear | 0.46 | 0.3306 | 0.2191 | 0.1387 | 6.54 | |
| 2FI | 0.48 | 0.4574 | 0.1560 | 0.0518 | 7.20 | |
| Quadratic | 0.089 | 0.9853 | 0.9706 | 0.9172 | 0.63 | Suggested |
| Cubic | 0.072 | 0.9959 | 0.9809 | 0.4736 | 4.00 | Aliased |
| Model summary statistics for Red Pigment Yield | ||||||
| Linear | 0.37 | 0.3146 | 0.2004 | 0.1230 | 4.29 | |
| 2FI | 0.39 | 0.4328 | 0.1178 | 0.0127 | 4.82 | |
| Quadratic | 0.096 | 0.9734 | 0.9468 | 0.8469 | 0.75 | Suggested |
| Cubic | 0.058 | 0.9958 | 0.9806 | 0.402 | 2.92 | Aliased |
| Model summary statistics for Dry Biomass Weight | ||||||
| Linear | 1.49 | 0.3828 | 0.2799 | 0.1902 | 69.91 | |
| 2FI | 1.58 | 0.4820 | 0.1943 | −0.0036 | 86.64 | |
| Quadratic | 0.42 | 0.9714 | 0.9429 | 0.8445 | 13.42 | Suggested |
| Cubic | 0.18 | 0.9977 | 0.9894 | 0.9790 | 1.81 | Aliased |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatachalam, M.; Shum-Chéong-Sing, A.; Dufossé, L.; Fouillaud, M. Statistical Optimization of the Physico-Chemical Parameters for Pigment Production in Submerged Fermentation of Talaromyces albobiverticillius 30548. Microorganisms 2020, 8, 711. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8050711
Venkatachalam M, Shum-Chéong-Sing A, Dufossé L, Fouillaud M. Statistical Optimization of the Physico-Chemical Parameters for Pigment Production in Submerged Fermentation of Talaromyces albobiverticillius 30548. Microorganisms. 2020; 8(5):711. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8050711
Chicago/Turabian StyleVenkatachalam, Mekala, Alain Shum-Chéong-Sing, Laurent Dufossé, and Mireille Fouillaud. 2020. "Statistical Optimization of the Physico-Chemical Parameters for Pigment Production in Submerged Fermentation of Talaromyces albobiverticillius 30548" Microorganisms 8, no. 5: 711. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8050711