Biochemical and Molecular Characterization of a Thermostable Alkaline Metallo-Keratinase from Bacillus sp. Nnolim-K1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Keratin Substrate
2.2. Bacterial Isolate Selection
2.3. Bacterial Resuscitation and Fresh Inoculum Preparation
2.4. Keratinase Production
2.4.1. Keratinase Activity Assay and Quantitation of Total Protein
2.4.2. Determination of Thiol Concentration
2.5. Construction of Optimal Process Variable for Enhanced Keratinase Production
2.5.1. One Variable at a Time (OVAT) Method
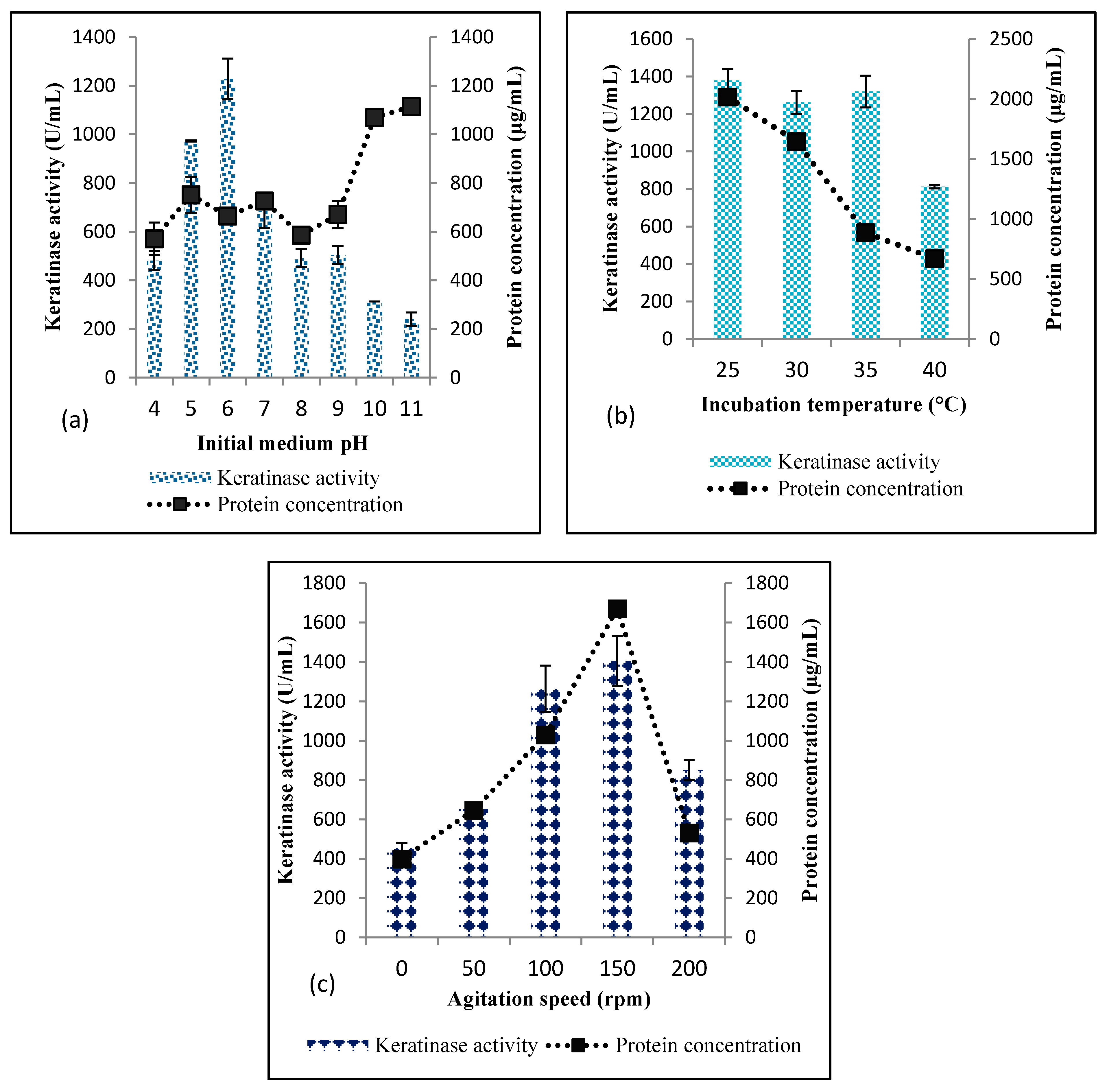
Effect of Initial Medium pH, Incubation Temperature and Agitation Speed
Effect of Extra Carbon and Nitrogen Supplementation
Effect of Chicken Feather Concentration
2.5.2. Statistical Optimization
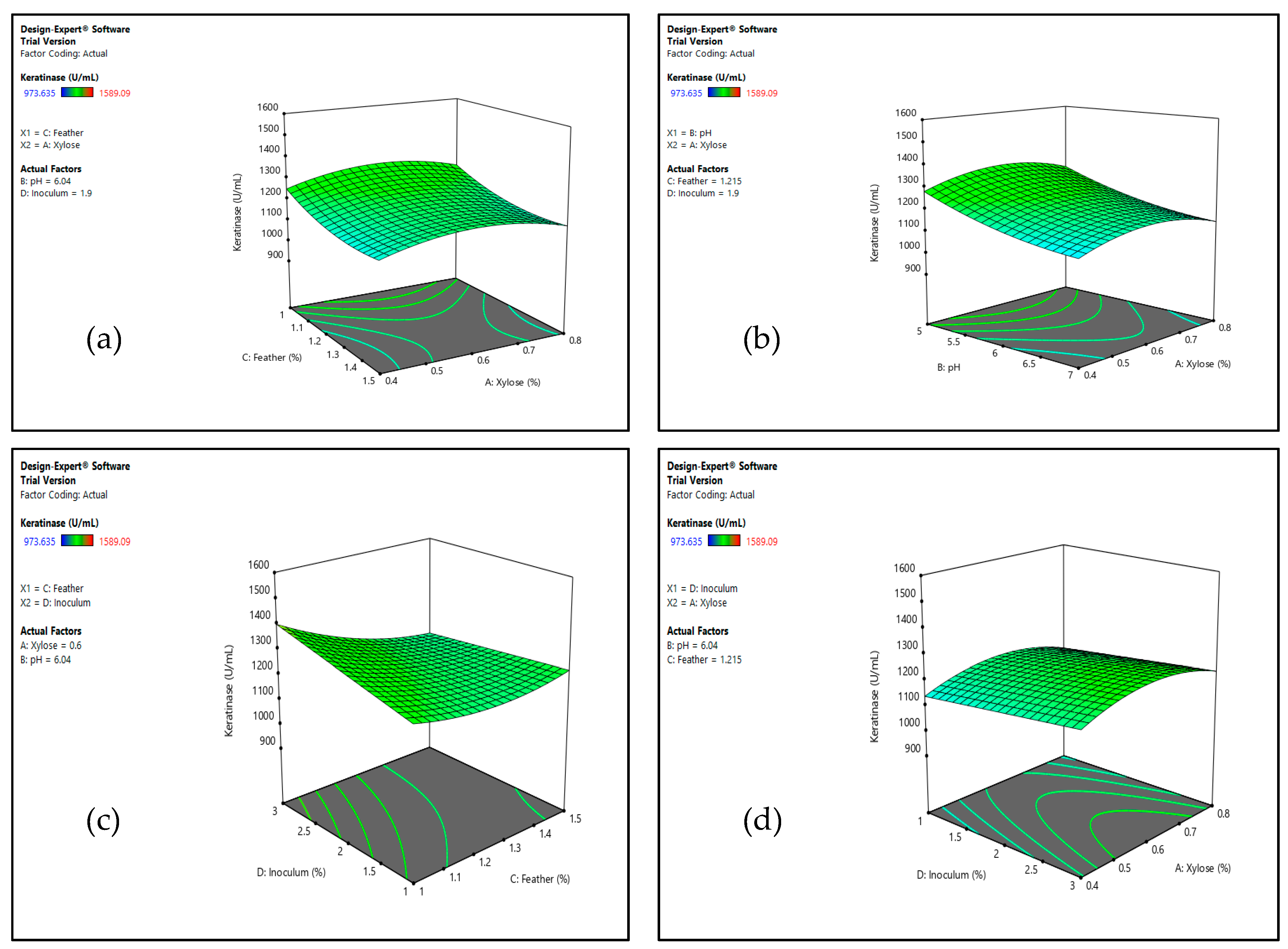
Experimental Design and Response Surface Methodology (RSM)
2.5.3. Time Course Profile of Keratinase Activity
2.6. Biochemical Characterization of Keratinase (KerBNK1)
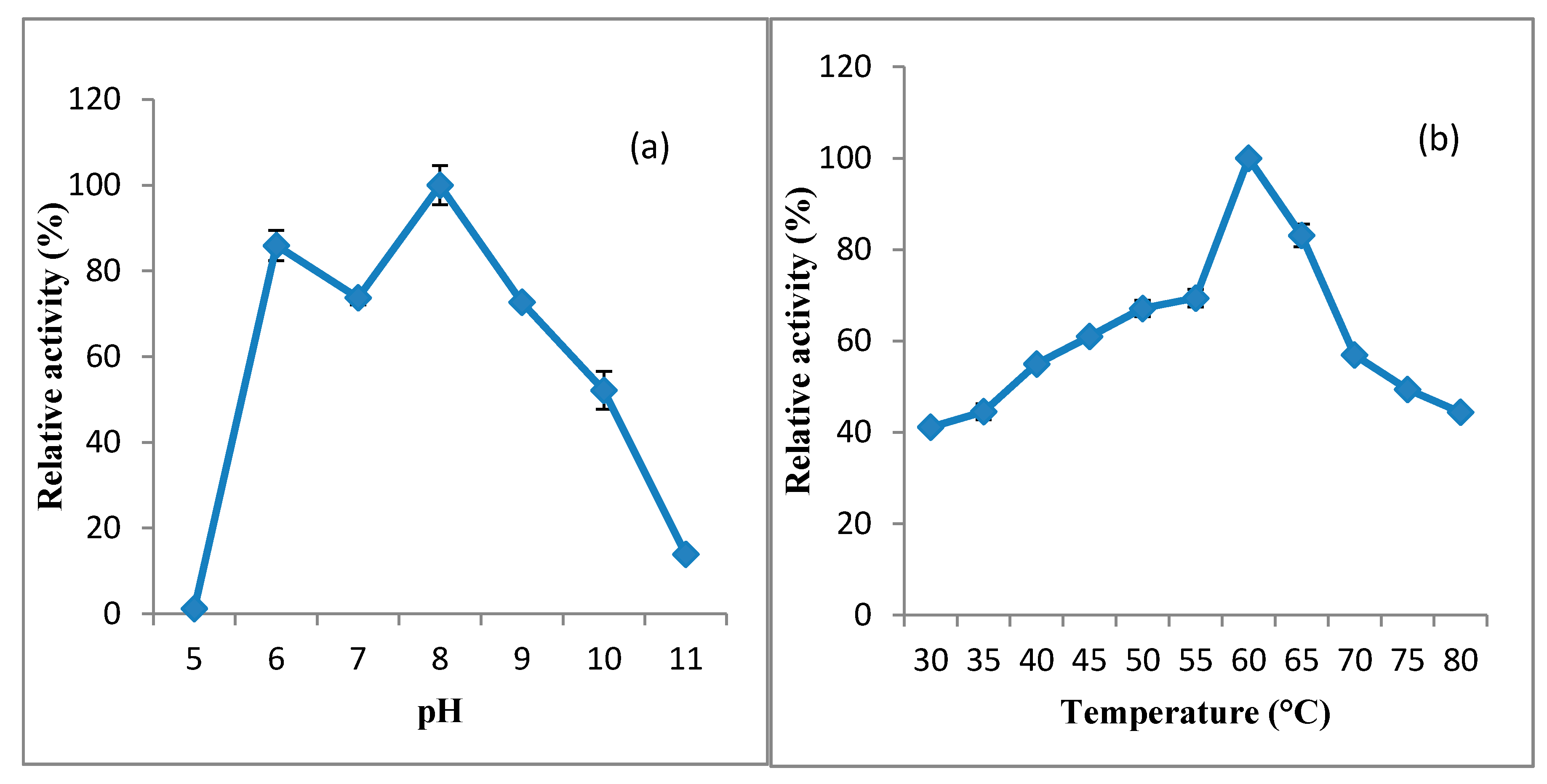
2.6.1. Effect of pH and Temperature on KerBNK1 Activity and Stability
2.6.2. Effect of Chemical Agents on KerBNK1 Stability
2.6.3. Effect of Metal Ions on KerBNK1 Stability
2.7. Molecular Characterization of KerBNK1
2.7.1. DNA Extraction
2.7.2. Keratinase Gene Amplification
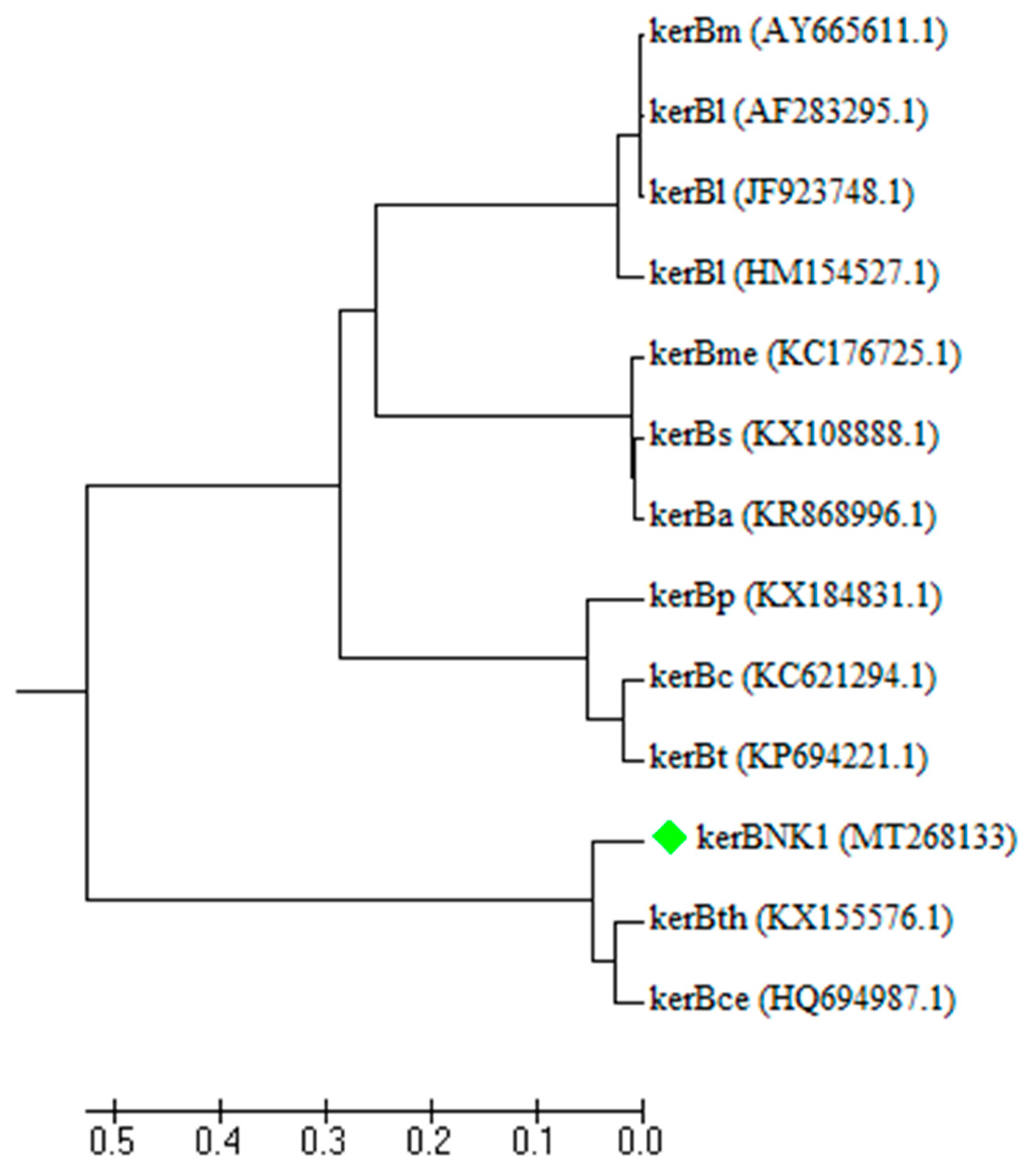
2.7.3. Sanger Sequencing and Phylogenetic Analyses
2.8. Evaluation of Laundry Detergent Impact on KerBNK1 Stability
2.9. Biodegradation of White and Melanized Feathers—Structural Evaluation
2.10. Statistical Analysis
3. Results and Discussion
3.1. Optimization of Keratinase Production by Bacillus sp. Nnolim-K1
3.1.1. Selection of Significant Physicochemical Parameters using OVAT
3.1.2. Optimization by Response Surface Methodology (RSM)
3.1.3. Time Course Profile of Keratinolytic Activity by Bacillus sp. Nnolim-K1
3.2. Biochemical Characterization of Keratinase (KerBNK1)
3.2.1. Effect of pH and Temperature on KerBNK1 Activity and Stability
3.2.2. Effect of Chemical Agents and Metal Ions on KerBNK1 Stability
3.3. Molecular Characterization of KerBNK1
3.4. Impact of Laundry Detergent on KerBNK1 Stability
3.5. Biodegradation of White and Melanized Feathers–Structural Study
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, B.; Yang, W.; McKittrick, J.; Meyers, M.A. Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog. Mater. Sci. 2016, 76, 229–318. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Zheng, X. Medium optimization for keratinase production in hair substrate by a new Bacillus subtilis KD-N2 using response surface methodology. J. Ind. Microbiol. Biotechnol. 2009, 36, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Onifade, A.A.; Al-Sane, N.A.; Al-Musallam, A.A.; Al-Zarban, S. A review: Potentials for biotechnological applications of keratin-degrading microorganisms and their enzymes for nutritional improvement of feathers and other keratins as livestock feed resources. Bioresour. Technol. 1998, 66, 1–11. [Google Scholar] [CrossRef]
- Stiborova, H.; Branska, B.; Vesela, T.; Lovecka, P.; Stranska, M.; Hajslova, J.; Jiru, M.; Patakova, P.; Demnerova, K. Transformation of raw feather waste into digestible peptides and amino acids. J. Chem. Technol. Biotechnol. 2016, 91, 1629–1637. [Google Scholar] [CrossRef]
- Jeong, J.H.; Lee, O.M.; Jeon, Y.D.; Kim, J.D.; Lee, N.R.; Lee, C.Y.; Son, H.J. Production of keratinolytic enzyme by a newly isolated feather-degrading Stenotrophomonas maltophilia that produces plant growth-promoting activity. Process Biochem. 2010a, 45, 1738–1745. [Google Scholar] [CrossRef]
- Williams, C.M.; Richter, C.S.; Mackenzie, J.M.; Shih, J.C. Isolation, identification, and characterization of a feather-degrading bacterium. Appl. Environ. Microbiol. 1990, 56, 1509–1515. [Google Scholar] [CrossRef] [Green Version]
- Daniels, G. The digestion of human hair keratin by Microsporum canis Bodin. Microbiology 1953, 8, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Gurav, R.G.; Tang, J.; Jadhav, J.P. Sulfitolytic and keratinolytic potential of Chryseobacterium sp. RBT revealed hydrolysis of melanin containing feathers. 3 Biotech 2016, 6, 145. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Singh, R. Hydrolyzing proficiency of keratinases in feather degradation. Ind. J. Microbiol. 2014, 54, 466–470. [Google Scholar] [CrossRef] [Green Version]
- Tatineni, R.; Doddapaneni, K.K.; Potumarthi, R.C.; Vellanki, R.N.; Kandathil, M.T.; Kolli, N.; Mangamoori, L.N. Purification and characterization of an alkaline keratinase from Streptomyces sp. Bioresour. Technol. 2008, 99, 1596–1602. [Google Scholar] [CrossRef]
- De Oliveira Martinez, J.P.; Cai, G.; Nachtschatt, M.; Navone, L.; Zhang, Z.; Robins, K.; Speight, R. Challenges and opportunities in identifying and characterising keratinases for value-added peptide production. Catalysts 2020, 10, 184. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Singh, H.; Anwar, S.; Chattopadhyay, A.; Tiwari, K.K.; Kaur, S.; Dhilon, G.S. Microbial keratinases: Industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 2017, 37, 476–491. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Rajput, R.; Sharma, R.; Gupta, N. Biotechnological applications and prospective market of microbial keratinases. Appl. Microbiol. Biotechnol. 2013, 97, 9931–9940. [Google Scholar] [CrossRef] [PubMed]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature—A new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusuf, I.; Ahmad, S.A.; Phang, L.Y.; Syed, M.A.; Shamaan, N.A.; Khalil, K.A.; Dahalan, F.A.; Shukor, M.Y. Keratinase production and biodegradation of polluted secondary chicken feather wastes by a newly isolated multi heavy metal tolerant bacterium-Alcaligenes sp. AQ05-001. J. Environ. Manage. 2016, 183, 182–195. [Google Scholar] [CrossRef]
- Tiwary, E.; Gupta, R. Medium optimization for a novel 58 kDa dimeric keratinase from Bacillus licheniformis ER-15: Biochemical characterization and application in feather degradation and dehairing of hides. Bioresour. Technol. 2010, 101, 6103–6110. [Google Scholar] [CrossRef]
- Paul, T.; Das, A.; Mandal, A.; Halder, S.K.; Jana, A.; Maity, C.; DasMohapatra, P.K.; Pati, B.R.; Mondal, K.C. An efficient cloth cleaning properties of a crude keratinase combined with detergent: Towards industrial viewpoint. J. Clean. Prod. 2014, 66, 672–684. [Google Scholar] [CrossRef]
- Reddy, M.R.; Reddy, K.S.; Chouhan, Y.R.; Bee, H.; Reddy, G. Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour. Technol. 2017, 243, 254–263. [Google Scholar] [CrossRef]
- Nnolim, N.E.; Okoh, A.I.; Nwodo, U.U. Bacillus sp. FPF-1 produced keratinase with high potential for chicken feather degradation. Molecules 2020, 25, 1505. [Google Scholar] [CrossRef] [Green Version]
- Nnolim, N.E.; Okoh, A.I.; Nwodo, U.U. Proteolytic bacteria isolated from agro-waste dumpsites produced keratinolytic enzymes. Biotechnol. Rep. 2020, 27, e00483. [Google Scholar] [CrossRef]
- Jaouadi, B.; Abdelmalek, B.; Fodil, D.; Ferradji, F.Z.; Rekik, H.; Zaraî, N.; Bejar, S. Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 2010, 101, 8361–8369. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Maugeri, T.L.; Carbone, M.; Fera, M.T.; Gugliandolo, C. Detection and differentiation of Vibrio vulnificus in seawater and plankton of a coastal zone of the Mediterranean Sea. Res. Microbiol. 2006, 157, 194–200. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Kim, J.M.; Lim, W.J.; Suh, H.J. Feather-degrading Bacillus species from poultry waste. Process Biochem. 2001, 37, 287–291. [Google Scholar] [CrossRef]
- Gupta, R.; Ramnani, P. Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechnol. 2006, 70, 21. [Google Scholar] [CrossRef]
- Daroit, D.J.; Brandelli, A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2014, 34, 372–384. [Google Scholar] [CrossRef]
- Gurav, R.G.; Jadhav, J.P. Biodegradation of keratinous waste by Chryseobacterium sp. RBT isolated from soil contaminated with poultry waste. J. Basic Microbiol. 2013, 53, 128–135. [Google Scholar] [CrossRef]
- Nam, G.W.; Lee, D.W.; Lee, H.S.; Lee, N.J.; Kim, B.C.; Choe, E.A.; Hwang, J.K.; Suhartono, M.T.; Pyun, Y.R. Native-feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch. Microbiol. 2002, 178, 538–547. [Google Scholar] [CrossRef]
- Hossain, M.S.; Azad, A.K.; Sayem, S.A.; Mostafa, G.; Hoq, M.M. Production and partial characterization of feather-degrading keratinolytic serine protease from Bacillus licheniformis MZK-3. J. Biol. Sci. 2007, 7, 599–606. [Google Scholar]
- Bach, E.; Daroit, D.J.; Corrêa, A.P.F.; Brandelli, A. Production and properties of keratinolytic proteases from three novel Gram-negative feather-degrading bacteria isolated from Brazilian soils. Biodegradation 2011, 22, 1191. [Google Scholar] [CrossRef]
- Kainoor, P.S.; Naik, G.R. Production and characterization of feather degrading keratinase from Bacillus sp. JB 99. Ind. J. Biotechnol. 2010, 9, 384–390. [Google Scholar]
- Kumar, E.V.; Srijana, M.; Chaitanya, K.; Reddy, Y.; Reddy, G. Biodegradation of poultry feathers by a novel bacterial isolate Bacillus altitudinis GVC11. Ind. J. Biotechnol. 2011, 10, 502–507. [Google Scholar]
- Li, Y.; Liu, X.; Zhang, L.; Ding, Z.; Xu, S.; Gu, Z.; Shi, G. Transcriptional Changes in the Xylose Operon in Bacillus licheniformis and Their Use in Fermentation Optimization. Int. J. Mol. Sci. 2019, 20, 4615. [Google Scholar] [CrossRef] [Green Version]
- Bose, A.; Pathan, S.; Pathak, K.; Keharia, H. Keratinolytic protease production by Bacillus amyloliquefaciens 6B using feather meal as substrate and application of feather hydrolysate as organic nitrogen input for agricultural soil. Waste Biomass Valor. 2014, 5, 595–605. [Google Scholar] [CrossRef]
- Brandelli, A.; Daroit, D.J.; Riffel, A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010, 85, 1735–1750. [Google Scholar] [CrossRef]
- Fakhfakh, N.; Kanoun, S.; Manni, L.; Nasri, M. Production and biochemical and molecular characterization of a keratinolytic serine protease from chicken feather-degrading Bacillus licheniformis RPk. Can. J. Microbiol. 2009, 55, 427–436. [Google Scholar] [CrossRef]
- Park, G.T.; Son, H.J. Keratinolytic activity of Bacillus megaterium F7-1, a feather-degrading mesophilic bacterium. Microbiol. Res. 2009, 164, 478–485. [Google Scholar] [CrossRef]
- Demir, T.; Hameş, E.E.; Öncel, S.S.; Vardar-Sukan, F. An optimization approach to scale up keratinase production by Streptomyces sp. 2M21 by utilizing chicken feather. Int. Biodeterior. Biodegrad. 2015, 103, 134–140. [Google Scholar] [CrossRef]
- Bhari, R.; Kaur, M.; Singh, R.S.; Pandey, A.; Larroche, C. Bioconversion of chicken feathers by Bacillus aerius NSMk2: A potential approach in poultry waste management. Bioresour. Technol. Rep. 2018, 3, 224–230. [Google Scholar] [CrossRef]
- Jeong, J.H.; Park, K.H.; Oh, D.J.; Hwang, D.Y.; Kim, H.S.; Lee, C.Y.; Son, H.J. Keratinolytic enzyme-mediated biodegradation of recalcitrant feather by a newly isolated Xanthomonas sp. P5. Polym. Degrad. Stab. 2010, 95, 1969–1977. [Google Scholar] [CrossRef]
- Rajput, R.; Gupta, R. Thermostable keratinase from Bacillus pumilus KS12: Production, chitin crosslinking and degradation of Sup35NM aggregates. Bioresour. Technol. 2013, 133, 118–126. [Google Scholar] [CrossRef]
- Ramnani, P.; Singh, R.; Gupta, R. Keratinolytic potential of Bacillus licheniformis RG1: Structural and biochemical mechanism of feather degradation. Can. J. Microbiol. 2005, 51, 191–196. [Google Scholar] [CrossRef]
- Gupta, S.; Nigam, A.; Singh, R. Purification and characterization of a Bacillus subtilis keratinase and its prospective application in feed industry. Acta Biol. Szeged. 2015, 59, 197–204. [Google Scholar]
- Prakash, P.; Jayalakshmi, S.K.; Sreeramulu, K. Purification and characterization of extreme alkaline, thermostable keratinase, and keratin disulfide reductase produced by Bacillus halodurans PPKS-2. Appl. Microbiol. Biotechnol. 2010, 87, 625–633. [Google Scholar] [CrossRef]
- Pawar, V.A.; Prajapati, A.S.; Akhani, R.C.; Patel, D.H.; Subramanian, R.B. Molecular and biochemical characterization of a thermostable keratinase from Bacillus altitudinis RBDV1. 3 Biotech 2018, 8, 107. [Google Scholar] [CrossRef]
- Gegeckas, A.; Šimkutė, A.; Gudiukaitė, R.; Čitavičius, D.J. Characterization and application of keratinolytic paptidases from Bacillus spp. Int. J. Biol. Macromol. 2018, 113, 1206–1213. [Google Scholar] [CrossRef]
- Riffel, A.; Lucas, F.; Heeb, P.; Brandelli, A. Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch. Microbiol. 2003, 179, 258–265. [Google Scholar] [CrossRef]
- Thys, R.C.S.; Lucas, F.S.; Riffel, A.; Heeb, P.; Brandelli, A. Characterization of a protease of a feather-degrading Microbacterium species. Lett. Appl. Microbiol. 2004, 39, 181–186. [Google Scholar] [CrossRef]
- Bressollier, P.; Letourneau, F.; Urdaci, M.; Verneuil, B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl. Environ. Microbiol. 1999, 65, 2570–2576. [Google Scholar] [CrossRef] [Green Version]
- Fang, Z.; Zhang, J.; Liu, B.; Du, G.; Chen, J. Biochemical characterization of three keratinolytic enzymes from Stenotrophomonas maltophilia BBE11-1 for biodegrading keratin wastes. Int. Biodeterior Biodegrad. 2013, 82, 166–172. [Google Scholar] [CrossRef]
- Mu, T.; Sun, H.; Zhang, M.; Wang, C. Sweet Potato Processing Technology; China Science Publishing and Media Ltd Academic Press: London, UK, 2017. [Google Scholar] [CrossRef]
- Dutta, B.; Banerjee, A.; Chakraborty, P.; Bandopadhyay, R. In silico studies on bacterial xylanase enzyme: Structural and functional insight. J. Genet. Eng. Biotechnol. 2018, 16, 749–756. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar]
- Jaouadi, B.; Ellouz-Chaabouni, S.; Ali, M.B.; Messaoud, E.B.; Naili, B.; Dhouib, A.; Bejar, S. Excellent laundry detergent compatibility and high dehairing ability of the Bacillus pumilus CBS alkaline proteinase (SAPB). Biotechnol. Bioprocess Eng. 2009, 14, 503–512. [Google Scholar] [CrossRef]
- Navone, L.; Speight, R. Understanding the dynamics of keratin weakening and hydrolysis by proteases. PLoS ONE 2018, 13, e0202608. [Google Scholar] [CrossRef]






| Code Value | Independent Variables | Range Coding Level | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| A(X1) | Xylose (%; w/v) | 0.4 | 0.6 | 0.8 |
| B(X2) | pH | 5.0 | 6.0 | 7.0 |
| C(X3) | Feather (%; w/v) | 1.0 | 1.25 | 1.5 |
| D(X4) | Inoculum (%; v/v) | 1.0 | 2.0 | 3.0 |
| Run | Independent Variable | Response: Keratinase Activity (U/mL) | ||||
|---|---|---|---|---|---|---|
| A: Xylose (%) | B: pH | C: Feather (%) | D: Inoculum (%) | Experimental | Predicted | |
| 1 | 0.6(0) | 7(+1) | 1.25(0) | 2(0) | 1181.82 | 1180.66 |
| 2 | 0.6(0) | 6(0) | 1.25(0) | 2(0) | 1280.91 | 1232.85 |
| 3 | 0.8(+1) | 5(−1) | 1(−1) | 1(−1) | 1320.91 | 1313.14 |
| 4 | 0.6(0) | 6(0) | 1.25(0) | 1(−1) | 1217.27 | 1209.21 |
| 5 | 0.8(+1) | 5(−1) | 1(−1) | 3(+1) | 1589.09 | 1460.64 |
| 6 | 0.6(0) | 5(−1) | 1.25(0) | 2(0) | 1367.27 | 1344.30 |
| 7 | 0.4(−1) | 6(0) | 1.25(0) | 2(0) | 1219.09 | 1162.38 |
| 8 | 0.8(+1) | 5(−1) | 1.5(+1) | 3(+1) | 1332.73 | 1249.40 |
| 9 | 0.8(+1) | 6(0) | 1.25(0) | 2(0) | 1146.36 | 1178.94 |
| 10 | 0.8(+1) | 7(+1) | 1(−1) | 3(+1) | 1217.27 | 1297.00 |
| 11 | 0.8(+1) | 7(+1) | 1.5(+1) | 3(+1) | 1145.45 | 1085.76 |
| 12 | 0.6(0) | 6(0) | 1.25(0) | 2(0) | 1369.09 | 1232.85 |
| 13 | 0.4(−1) | 5(−1) | 1(−1) | 1(−1) | 1253.64 | 1296.57 |
| 14 | 0.6(0) | 6(0) | 1.25(0) | 2(0) | 1318.18 | 1232.85 |
| 15 | 0.8(+1) | 7(+1) | 1(−1) | 1(−1) | 1130.91 | 1149.50 |
| 16 | 0.6(0) | 6(0) | 1.25(0) | 2(0) | 1164.54 | 1232.85 |
| 17 | 0.4(−1) | 7(+1) | 1(−1) | 1(−1) | 1146.36 | 1132.93 |
| 18 | 0.4(−1) | 5(−1) | 1.5(+1) | 3(+1) | 1213.64 | 1232.83 |
| 19 | 0.8(+1) | 7(+1) | 1.5(+1) | 1(−1) | 1145.45 | 1138.72 |
| 20 | 0.4(−1) | 7(+1) | 1(−1) | 3(+1) | 1369.09 | 1280.43 |
| 21 | 0.6(0) | 6(0) | 1.5(+1) | 2(0) | 1253.64 | 1218.34 |
| 22 | 0.6(0) | 6(0) | 1(−1) | 2(0) | 1318.18 | 1329.35 |
| 23 | 0.6(0) | 6(0) | 1.25(0) | 2(0) | 1312.73 | 1232.85 |
| 24 | 0.4(−1) | 5(−1) | 1.5(+1) | 1(−1) | 1346.36 | 1285.79 |
| 25 | 0.4(−1) | 5(−1) | 1(−1) | 3(+1) | 1358.18 | 1444.07 |
| 26 | 0.6(0) | 6(0) | 1.25(0) | 2(0) | 1070.91 | 1232.85 |
| 27 | 0.4(−1) | 7(+1) | 1.5(+1) | 1(−1) | 1146.36 | 1122.15 |
| 28 | 0.8(+1) | 5(−1) | 1.5(+1) | 1(−1) | 1147.27 | 1302.35 |
| 29 | 0.6(0) | 6(0) | 1.25(0) | 3(+1) | 1080.91 | 1256.49 |
| 30 | 0.4(−1) | 7(+1) | 1.5(+1) | 3(+1) | 973.64 | 1069.20 |
| Chemical Agent | Concentration | Residual Activity (%) |
|---|---|---|
| None | - | 100 ± 3.92 |
| EDTA | 5 mM | 0 ± 0 |
| 1,10-phenanthroline | 5 mM | 0 ± 0 |
| PMSF | 5 mM | 96 ± 2.62 |
| Sodium thioglycolate | 5 mM | 76 ± 0.51 |
| Dithiothreitol | 5 mM | 92 ± 0.29 |
| Sodium sulfite | 5 mM | 87 ± 0.65 |
| 2-Mercaptoethanol | 5 mM | 102 ± 4.79 |
| Hydrogen peroxide | 1% (v/v) | 91 ± 2.69 |
| Acetonitrile | 1% (v/v) | 91 ± 0.29 |
| Propan-2-ol | 1% (v/v) | 75 ±0.95 |
| Sodium dodecyl sulfate | 5 mM | 84 ± 1.53 |
| Triton X-100 | 1% (v/v) | 106 ± 2.54 |
| Tween-80 | 1% (v/v) | 119 ± 5.59 |
| Metal Ion | Concentration (mM) | Residual Activity (%) |
|---|---|---|
| None | - | 100 ± 3.92 |
| Li+ | 5 | 96 ± 1.96 |
| Na+ | 5 | 76 ± 4.87 |
| K+ | 5 | 94 ± 1.35 |
| Ag+ | 5 | 96 ± 1.59 |
| Ca2+ | 5 | 116 ± 5.16 |
| Mg2+ | 5 | 97 ± 3.56 |
| Mn2+ | 5 | 91 ± 1.38 |
| Zn2+ | 5 | 72 ± 0.95 |
| Cu2+ | 5 | 77 ± 5.09 |
| Fe2+ | 5 | 86 ± 2.18 |
| Hg2+ | 5 | 82 ± 0.07 |
| Ba2+ | 5 | 70 ± 3.19 |
| Co2+ | 5 | 66 ± 5.67 |
| Al3+ | 5 | 89 ±2.83 |
| Fe3+ | 5 | 77 ± 0.65 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nnolim, N.E.; Mpaka, L.; Okoh, A.I.; Nwodo, U.U. Biochemical and Molecular Characterization of a Thermostable Alkaline Metallo-Keratinase from Bacillus sp. Nnolim-K1. Microorganisms 2020, 8, 1304. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8091304
Nnolim NE, Mpaka L, Okoh AI, Nwodo UU. Biochemical and Molecular Characterization of a Thermostable Alkaline Metallo-Keratinase from Bacillus sp. Nnolim-K1. Microorganisms. 2020; 8(9):1304. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8091304
Chicago/Turabian StyleNnolim, Nonso E., Lindelwa Mpaka, Anthony I. Okoh, and Uchechukwu U. Nwodo. 2020. "Biochemical and Molecular Characterization of a Thermostable Alkaline Metallo-Keratinase from Bacillus sp. Nnolim-K1" Microorganisms 8, no. 9: 1304. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8091304





