Syngas as Electron Donor for Sulfate and Thiosulfate Reducing Haloalkaliphilic Microorganisms in a Gas-Lift Bioreactor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioreactor Set-Up
2.2. Inoculum
2.3. Medium
2.4. Experimental Design
2.5. Batch Experiments
2.6. Analytical Procedures
2.7. Scanning Electron Microscopy
2.8. DNA Isolation and Bacteria Community Profiling
2.9. Calculations
3. Results and Discussion
3.1. Bioreactor Performance
3.2. Inhibition by CO
3.3. Adaptation to CO
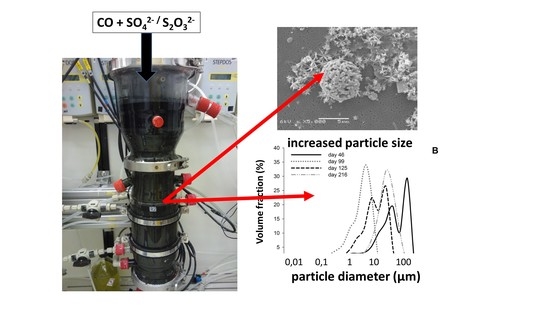
3.4. Biomass Aggregation
3.5. Application in Gas Biodesulfurization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Nomenclature
| Cx | Biomass concentration (g L−1) |
| Ctx | Total biomass concentration (g L−1) |
| Mx | Biomass molecular weight (g mol−1) |
| Vr | Bioreactor volume (L) |
| Q | Flow rate (L d−1) |
| Cs | Sulfate or thiosulfate concentration based on S molar content (mmolS L−1) |
| CTotalN | Total nitrogen concentration (mol L−1) |
| rvs | Sulfidogenic volumetric activity (mmolS Lr−1 d−1) |
| rvp | Formate, acetate or methane production volumetric activity (mmol Lr−1 d−1) |
| Subscript | |
| a | Attached biomass |
| x | Biomass |
| out | Effluent |
| in | Influent |
| p | Products |
| r | Reactor |
| s | Sulfur |
| sp | Suspended biomass |
| tx | Total biomass |
| TotalN | Total nitrogen |
References
- Janssen, A.J.H.; Lens, P.N.L.; Stams, A.J.M.; Plugge, C.M.; Sorokin, D.Y.; Muyzer, G.; Dijkman, H.; Van Zessen, E.; Luimes, P.; Buisman, C.J.N. Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci. Total Environ. 2009, 407, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- De Rink, R.; Klok, J.B.M.; Van Heeringen, G.J.; Sorokin, D.Y.; Ter Heijne, A.; Zeijlmaker, R.; Mos, Y.M.; de Wilde, V.; Keesman, K.J.; Buisman, C.J.N. Increasing the selectivity for sulfur formation in biological gas desulfurization. Environ. Sci. Technol. 2019, 53, 4519–4527. [Google Scholar] [CrossRef] [PubMed]
- Klok, J.B.M.; de Graaff, M.; van den Bosch, P.L.F.; Boelee, N.C.; Keesman, K.J.; Janssen, A.J.H. A physiologically based kinetic model for bacterial sulfide oxidation. Water Res. 2013, 47, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Roman, P.; Bijmans, M.F.M.; Janssen, A.J.H. Influence of methanethiol on biological sulphide oxidation in gas treatment system. Environ. Technol. 2016, 37, 1693–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Z.; Tianwei, H.; Mackey, H.R.; van Loosdrecht, M.C.M.; Guanghao, C. Recent advances in dissimilatory sulfate reduction. Water Res. 2019, 150, 162–181. [Google Scholar] [CrossRef]
- Mu, T.; Yang, M.; Xing, J. Deep and high-efficiency removal of sulfate through a coupling system with sulfate-reducing and sulfur-oxidizing capacity under haloalkaliphilic condition. Bioprocess Biosyst. Eng. 2020, 43, 1009–1015. [Google Scholar] [CrossRef]
- Sousa, J.A.B.; Plugge, C.M.; Stams, A.J.M.; Bijmans, M.F.M. Sulfate reduction in a hydrogen fed bioreactor operated at haloalkaline conditions. Water Res. 2015, 68, 67–76. [Google Scholar] [CrossRef]
- Sousa, J.A.B.; Bijmans, M.F.M.; Stams, A.J.M.; Plugge, C.M. Thiosulfate conversion to sulfide by a haloalkaliphilic microbial community in a bioreactor fed with H2 gas. Environ. Sci. Technol. 2017, 51, 914–923. [Google Scholar] [CrossRef]
- Zhou, J.; Xing, J. Effect of electron donors on the performance of haloalkaliphilic sulfate-reducing bioreactors for flue gas treatment and microbial degradation patterns related to sulfate reduction of different electron donors. Biochem. Eng. J. 2015, 96, 14–22. [Google Scholar] [CrossRef]
- Van Houten, B.H.G.W.; van Doesburg, W.; Dijkman, H.; Copini, C.; Smidt, H.; Stams, A.J.M. Long-term performance and microbial community analysis of a full-scale synthesis gas fed reactor treating sulfate- and zinc-rich wastewater. Appl. Microbiol. Biotechnol. 2009, 84, 555–563. [Google Scholar] [CrossRef] [Green Version]
- Diender, M.; Stams, A.J.M.; Sousa, D.Z. Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol. 2015, 6, 1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parshina, S.N.; Sipma, J.; Henstra, A.M.; Stams, A.J.M. Carbon monoxide as an electron donor for the biological reduction of sulphate. Int. J. Microbiol. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Jeoung, J.H.; Fesseler, J.; Goetzl, S.; Dobbek, H. Carbon monoxide. Toxic gas and fuel for anaerobes and aerobes: Carbon monoxide dehydrogenases. In Metal Ions in Life Sciences; Kroneck, P.M.H., Sosa Torres, M.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 37–69. [Google Scholar] [CrossRef]
- Lubitz, W.; Hideaki, O.; Rudiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.W.W. The structure and mechanism of iron-hydrogenases. Biochim. Biophys. Acta Bioenerg. 1990, 1020, 115–145. [Google Scholar] [CrossRef]
- De Lacey, A.L.; Fernández, V.M.; Rousset, M.; Cammack, R. Activation and inactivation of hydrogenase function and the catalytic cycle: Spectroelectrochemical studies. Chem. Rev. 2007, 107, 4304–4330. [Google Scholar] [CrossRef]
- Takors, R.; Kopf, M.; Mampel, J.; Bluemke, W.; Blombach, B.; Eikmanns, B.; Bengelsdorf, F.R.; Weuster-Botz, D.; Dürre, P. Using gas mixtures of CO, CO2 and H2 as microbial substrates: The do’s and don’ts of successful technology transfer from laboratory to production scale. Microb. Biotechnol. 2018, 11, 606–625. [Google Scholar] [CrossRef]
- Sipma, J.; Lens, P.N.L.; Stams, A.J.M.; Lettinga, G. Carbon monoxide conversion by anaerobic bioreactor sludges. FEMS Microbiol. Ecol. 2003, 44, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Robb, F.T.; Techtman, S.M. Life on the fringe: Microbial adaptation to growth on carbon monoxide. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Parshina, S.N.; Kijlstra, S.; Henstra, A.M.; Sipma, J.; Plugge, C.M.; Stams, A.J.M. Carbon monoxide conversion by thermophilic sulfate-reducing bacteria in pure culture and in co-culture with Carboxydothermus hydrogenoformans. Appl. Microbiol. Biotechnol. 2005, 68, 390–396. [Google Scholar] [CrossRef]
- Parshina, S.N.; Sipma, J.; Nakashimada, Y.; Henstra, A.M.; Smidt, H.; Lysenko, A.M.; Lens, P.N.L.; Lettinga, G.; Stams, A.J.M. Desulfotomaculum carboxydivorans sp. nov., a novel sulfate-reducing bacterium capable of growth at 100% CO. Int. J. Syst. Evol. Microbiol. 2005, 55, 2159–2165. [Google Scholar] [CrossRef] [Green Version]
- Van Houten, R.T.; van der Spoel, H.; van Aelst, A.C.; Hulshoff Pol, L.W.; Lettinga, G. Biological sulfate reduction using synthesis gas as energy and carbon source. Biotechnol. Bioeng. 1996, 50, 136–144. [Google Scholar] [CrossRef]
- Sousa, J.A.B.; Bolgár, A.; Christel, S.; Dopson, M.; Bijmans, M.F.M.; Stams, A.J.M.; Plugge, C.M. Immobilization of sulfate/thiosulfate-reducing biomass on sand under haloalkaline conditions. Sci. Total Environ. 2020, 745, 141017. [Google Scholar] [CrossRef] [PubMed]
- Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Formation of methane by bacterial extracts. J. Biol. Chem. 1963, 238, 2882–2886. [Google Scholar] [PubMed]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugerth, L.W.; Wefer, H.A.; Lundin, S.; Jakobsson, H.E.; Lindberg, M.; Rodin, S.; Engstrand, L.; Andersson, A.F. DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl. Environ. Microbiol. 2014, 80, 5116–5123. [Google Scholar] [CrossRef] [Green Version]
- Lindh, M.V.; Figueroa, D.; Sjöstedt, J.; Baltar, F.; Lundin, D.; Andersson, A.; Legrand, C.; Pinhassi, J. Transplant experiments uncover Baltic Sea basin-specific responses in bacterioplankton community composition and metabolic activities. Front. Microbiol. 2015, 6, 223. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Robertson, C.E.; Harris, J.K.; Wagner, B.D.; Granger, D.; Browne, K.; Tatem, B.; Feazel, L.M.; Pace, N.R.; Frank, D.N. Explicet: Graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 2013, 29, 3100–3101. [Google Scholar] [CrossRef] [Green Version]
- Flamholz, A.; Noor, E.; Bar-Even, A.; Milo, R. eQuilibrator-the biochemical thermodynamics calculator. Nucleic Acids Res. 2012, 40, D770–D775. [Google Scholar] [CrossRef] [Green Version]
- Ferry, J.G. CO in methanogenesis. Ann. Microbiol. 2010, 60, 1–12. [Google Scholar] [CrossRef]
- Esquivel-Elizondo, S.; Delgado, A.G.; Rittmann, B.E.; Krajmalnik-Brown, R. The effects of CO2 and H2 on CO metabolism by pure and mixed microbial cultures. Biotechnol. Biofuels 2017, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Bertsch, J.; Müller, V. CO metabolism in the acetogen Acetobacterium woodii. Appl. Environ. Microbiol. 2015, 81, 5949–5956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, L.; Fuchs, G.; Thauer, R.K.; Zeikus, J.G. Carbon monoxide oxidation by methanogenic bacteria. J. Bacteriol. 1977, 132, 118–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.W.W. The metabolism of hydrogen by extremely thermophilic, sulfur-dependent bacteria. FEMS Microbiol. Rev. 1990, 6, 219–237. [Google Scholar] [CrossRef]
- Fox, J.D.; Kerby, R.L.; Roberts, G.P.; Ludden, P.W. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J. Bacteriol. 1996, 178, 1515–1524. [Google Scholar] [CrossRef] [Green Version]
- Ceccaldi, P.; Schuchmann, K.; Muller, V.; Elliott, S.J. The hydrogen dependent CO2 reductase: The first completely CO tolerant FeFe-hydrogenase. Energy Environ. Sci. 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Techtmann, S.M.; Colman, A.S.; Robb, F.T. “That which does not kill us only makes us stronger”: The role of carbon monoxide in thermophilic microbial consortia. Environ. Microbiol. 2009, 11, 1027–1037. [Google Scholar] [CrossRef]
- Henstra, A.M.; Dijkema, C.; Stams, A.J.M. Archaeoglobus fulgidus couples CO oxidation to sulfate reduction and acetogenesis with transient formate accumulation. Environ. Microbiol. 2007, 9, 1836–1841. [Google Scholar] [CrossRef]
- Köpke, M.; Held, C.; Hujer, S.; Liesegang, H.; Wiezer, A.; Wollherr, A.; Ehrenreich, A.; Liebl, W.; Gottschalk, G.; Dürre, P. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. USA 2010, 107, 15305. [Google Scholar] [CrossRef] [Green Version]
- Rother, M.; Metcalf, W.W. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: An unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. USA 2004, 101, 16929–16934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhilina, T.N.; Zavarzina, D.G.; Panteleeva, A.N.; Osipov, G.A.; Kostrikina, N.A.; Tourova, T.P.; Zavarzin, G.A. Fuchsiella alkaliacetigena gen. nov., sp. nov., an alkaliphilic, lithoautotrophic homoacetogen from a soda lake. Int. J. Syst. Evol. Microbiol. 2012, 62, 1666–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhilina, T.N.; Kuznetsov, B.B.; Zavarzina, D.G.; Detkova, E.N.; Patutina, E.O. Fuchsiella ferrireducens sp. nov., a novel haloalkaliphilic, lithoautotrophic homoacetogen capable of iron reduction, and emendation of the description of the genus Fuchsiella. Int. J. Syst. Evol. Microbiol. 2015, 65, 2432–2440. [Google Scholar] [CrossRef]
- Mu, T.; Xing, J.; Yang, M. Sulfate reduction by a haloalkaliphilic bench-scale sulfate-reducing bioreactor and its bacterial communities at different depths. Biochem. Eng. J. 2019, 147, 100–109. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Detkova, E.N.; Muyzer, G. Sulfur-dependent respiration under extremely haloalkaline conditions in soda lake “acetogens” and the description of Natroniella sulfidigena sp. nov. FEMS Microbiol. Lett. 2011, 319, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sipma, J.; Lettinga, G.; Stams, A.J.M.; Lens, P.N.L. Hydrogenogenic CO conversion in a moderately thermophilic (55 °C) sulfate-fed gas lift reactor: Competition for CO-derived H2. Biotechnol. Prog. 2006, 22, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Sinharoy, A.; Baskaran, D.; Pakshirajan, K. A novel carbon monoxide fed moving bed biofilm reactor for sulfate rich wastewater treatment. J. Environ. Manag. 2019, 249, 109402. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Abbas, B.; Suhacheva, M.V.; Muyzer, G. Desulfonatronovibrio halophilus sp. nov., a novel moderately halophilic sulfate-reducing bacterium from hypersaline chloride-sulfate lakes in Central Asia. Extremophiles 2012, 16, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Ismail, S.B.; Gonzalez, P.; Jeison, D.; van Lier, J.B. Effects of high salinity wastewater on methanogenic sludge bed systems. Water Sci. Technol. 2008, 58, 1963–1970. [Google Scholar] [CrossRef]
- Otto, K.; Elwing, H.; Hermansson, M. Effect of ionic strength on initial interactions of Escherichia coli with surfaces, studied on-line by a novel quartz crystal microbalance technique. J. Bacteriol. 1999, 181, 5210–5218. [Google Scholar] [CrossRef] [Green Version]
- Sousa, J.A.B.; Sorokin, D.Y.; Bijmans, M.F.M.; Plugge, C.M.; Stams, A.J.M. Ecology and application of haloalkaliphilic anaerobic microbial communities. Appl. Microbiol. Biotechnol. 2015, 99, 9331–9337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, G.; Kaewpanha, M.; Hao, X.; Abudula, A. Catalytic steam reforming of biomass tar: Prospects and challenges. Renew. Sustain. Energy Rev. 2016, 58, 450–461. [Google Scholar] [CrossRef] [Green Version]
- Voldsund, M.; Jordal, K.; Anantharaman, R. Hydrogen production with CO2 capture. Int. J. Hydrog. Energy 2016, 41, 4969–4992. [Google Scholar] [CrossRef]
- Van den Bosch, P.L.F. Biological Sulfide Oxidation by Natron-Alkaliphilic Bacteria Application in Gas Desulfurization. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2008. [Google Scholar]








| Reaction | ΔG’ Starting (kJ mol−1) | ΔG’ Continuous Operation (kJ mol−1) | Equation No. |
|---|---|---|---|
| CO + 2H2O → HCO3− + H2 + H+ | −22.7 (±10.5) | −23.1 (±10.5) | (1) |
| 4CO + 4H2O → C2H3O2− + 2HCO3− + 3H+ | −199.6 (±26.6) | −182.9 (±26.6) | (2) |
| 4H2 + 2HCO3− + H+ → C2H3O2− + 4H2O | −108.9 (±26.9) | −90.5 (±26.9) | (3) |
| 4CO + SO42− + 4H2O → 4HCO3− + HS− + 3H+ | −240.6 (±34.3) | −218.8 (±34.3) | (4) |
| 4H2 + SO42− + H+ → HS− + 4H2O | −150.0 (±25.0) | −126.4 (±25.0) | (5) |
| CO + H2O → HCO2− + H+ | −38.0 (±6.5) | −32.3 (±6.5) | (6) |
| H2 + HCO3− → HCO2− + H2O | −15.2 (±8.9) | −9.2 (±8.9) | (7) |
| 4CO + 5H2O → CH4 + 3HCO3− + 3H+ | −215.4 (±30.6) | −198 (±30.6) | (8) |
| 4H2 + HCO3− + H+ → CH4 + 3H2O | −124.4 (±25.1) | −105.6 (±25.1) | (9) |
| Period (Days) | Mode | |
|---|---|---|
| Start-up | 0–6 | Batch |
| 0% CO | 7–45 | Continuous |
| 5% CO | 46–123 | Continuous |
| 15% CO | 124–216 | Continuous |
| 60% CO spike * | 217–235 | Continuous |
| This Study | Sousa et al. 2020 | Zhou and Xing 2015 | Sousa et al. 2017 | |
|---|---|---|---|---|
| Reactor type | Gas lift with 3 phase separator | Gas lift with 3 phase separator | Anaerobic filter | Gas lift with 3 phase separator |
| e− acceptor | Sulfate/thiosulfate | Sulfate/thiosulfate | Sulfate | Thiosulfate |
| e− donor | H2/CO | H2 | Formate | H2 |
| pH | 9 | 9 | 9.5 | 9 |
| Na+ conc. (M) | 1.5 | 1.5 | 1 | 1.5 |
| Temperature (°C) | 35 | 35 | 37 | 35 |
| HRT (d) | 1 | 1 | 1 | 1.7 |
| Max loading rate (mmolS L−1 d−1) | 50 | 100 | 88.5 | 50 |
| Sulfidogenic rate (mmolS L−1 d−1) | 46.8 (±0.8) | 85.45 (±3) | 85.05 (±0.2) | 28.7 (±0.8) |
| Side products | Formate/Acetate/Methane | Formate/Acetate/Methane | Acetate | Formate |
| Biomass conc. (mg L−1) | 197 (±39) | 127 (±41) | N.D. | 14 (±2.2) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plugge, C.M.; Sousa, J.A.B.; Christel, S.; Dopson, M.; Bijmans, M.F.M.; Stams, A.J.M.; Diender, M. Syngas as Electron Donor for Sulfate and Thiosulfate Reducing Haloalkaliphilic Microorganisms in a Gas-Lift Bioreactor. Microorganisms 2020, 8, 1451. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8091451
Plugge CM, Sousa JAB, Christel S, Dopson M, Bijmans MFM, Stams AJM, Diender M. Syngas as Electron Donor for Sulfate and Thiosulfate Reducing Haloalkaliphilic Microorganisms in a Gas-Lift Bioreactor. Microorganisms. 2020; 8(9):1451. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8091451
Chicago/Turabian StylePlugge, Caroline M., João A. B. Sousa, Stephan Christel, Mark Dopson, Martijn F. M. Bijmans, Alfons J. M. Stams, and Martijn Diender. 2020. "Syngas as Electron Donor for Sulfate and Thiosulfate Reducing Haloalkaliphilic Microorganisms in a Gas-Lift Bioreactor" Microorganisms 8, no. 9: 1451. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8091451