Genetic Adaptations in Mudskipper and Tetrapod Give Insights into Their Convergent Water-to-Land Transition
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection, Alignment, and Trimming of Singleton Orthologues
2.2. Identification of Positive Selection Related to Water-to-Land Transition
2.3. Detection of Amino Acid Substitution under Positive Selection
2.4. Detection of Rapidly Evolving Gene Families
3. Results
3.1. Identifications and Classifications of Positively Selected Genes under Different Evolutionary Models Related to Water-to-Land Transition
3.2. Amino Acid Substitutions on the Functional Domains in Union PSGs
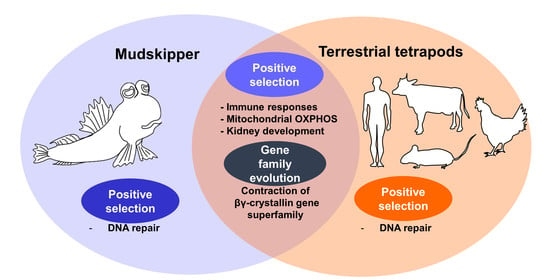
3.3. Enrichment of DNA Repair Processes on Tetrapod-Specific and Mudskipper-Specific PSGs
3.4. Convergent Contraction of βγ-Crystallin Gene Superfamily in Land-Adapting Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PSG | Positively selected gene |
| PSS | Positively selected site |
| BEB | Bayes empirical Bayes |
| TAAS | Target-specific amino acid substitution |
| EAASPS | Exclusive amino acid substitution under positive selection |
| FDR | False discovery rate |
References
- Daeschler, E.B.; Shubin, N.H.; Jenkins, F.A. A Devonian tetrapod-like fish and the evolution of the tetrapod body plan. Nat. Cell Biol. 2006, 440, 757–763. [Google Scholar] [CrossRef]
- Li, C.; Wu, X.-C.; Rieppel, O.; Wang, L.-T.; Zhao, L.-J. An ancestral turtle from the Late Triassic of southwestern China. Nat. Cell Biol. 2008, 456, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Hsia, C.C.W.; Schmitz, A.; Lambertz, M.; Perry, S.F.; Maina, J.N. Evolution of Air Breathing: Oxygen Homeostasis and the Transitions from Water to Land and Sky. Compr. Physiol. 2013, 3, 849–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayer, M.D.J.; Davenport, J. Amphibious fish: Why do they leave water? Rev. Fish Biol. Fish. 1991, 1, 159–181. [Google Scholar] [CrossRef]
- Graham, J.B.; Rosenblatt, R.H. Aerial Vision: Unique Adaptation in an Intertidal Fish. Science 1970, 168, 586–588. [Google Scholar] [CrossRef]
- Polgar, G.; Malavasi, S.; Cipolato, G.; Georgalas, V.; Clack, J.A.; Torricelli, P. Acoustic Communication at the Water’s Edge: Evolutionary Insights from a Mudskipper. PLoS ONE 2011, 6, e21434. [Google Scholar] [CrossRef] [Green Version]
- Kawano, S.M.; Blob, R.W. Propulsive forces of mudskipper fins and salamander limbs during terrestrial locomotion: Implicaions for the invasion of land. Integr. Comp. Biol. 2013, 53, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Pace, C.M.; Gibb, A.C. Mudskipper pectoral fin kinematics in aquatic and terrestrial environments. J. Exp. Biol. 2009, 212, 2279–2286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teal, J.M.; Carey, F.G. Skin Respiration and Oxygen Debt in the Mudskipper Periopthalmus sobrinus. Copeia 1967, 1967, 677. [Google Scholar] [CrossRef]
- Katayama, Y.; Sakamoto, T.; Saito, K.; Tsuchimochi, H.; Kaiya, H.; Watanabe, T.; Pearson, J.T.; Takei, Y. Drinking by amphibious fish: Convergent evolution of thirst mechanisms during vertebrate terrestrialization. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Peng, K.-W.; Chew, S.; Lim, C.; Kuah, S.; Kok, W.; Ip, Y. The mudskippers Periophthalmodon schlosseri and Boleoph-thalmus boddaerti can tolerate environmental NH 3 concentrations of 446 and 36µM, respectively. Fish Physiol. Biochem. 1998, 19, 59–69. [Google Scholar] [CrossRef]
- Sakamoto, T.; Yokota, S.; Ando, M. Rapid morphological oscillation of mitochondrion-rich cell in estuarine mudskipper following salinity changes. J. Exp. Zool. 2000, 286, 666–669. [Google Scholar] [CrossRef]
- Lee, H.J.; Martinez, C.A.; Hertzberg, K.J.; Hamilton, A.L.; Graham, J.B. Burrow air phase maintenance and respiration by the mudskipper Scartelaos histophorus (Gobiidae: Oxudercinae). J. Exp. Biol. 2005, 208, 169–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storz, J.F.; Natarajan, C.; Grouleff, M.K.; Vandewege, M.; Hoffmann, F.G.; You, X.; Venkatesh, B.; Fago, A. Oxygenation properties of hemoglobin and the evolutionary origins of isoform multiplicity in an amphibious air-breathing fish, the blue-spotted mudskipper (Boleophthalmus pectinirostris). J. Exp. Biol. 2020, 223, jeb217307. [Google Scholar] [CrossRef] [PubMed]
- Kutschera, U.; Elliott, J.M. Do mudskippers and lungfishes elucidate the early evolution of four-limbed vertebrates? Evol. Educ. Outreach 2013, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Jew, C.J.; Wegner, N.C.; Yanagitsuru, Y.; Tresguerres, M.; Graham, J.B. Atmospheric Oxygen Levels Affect Mudskipper Terrestrial Performance: Implications for Early Tetrapods. Integr. Comp. Biol. 2013, 53, 248–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niedźwiedzki, G.; Szrek, P.; Narkiewicz, K.; Narkiewicz, M.; Ahlberg, P.E. Tetrapod trackways from the early Middle Devonian period of Poland. Nat. Cell Biol. 2010, 463, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Clack, J.A. Gaining Ground: The Origin and Evolution of Tetrapods; Indiana University Press: Bloomington, Indiana, 2012. [Google Scholar]
- Graham, J.B.; Lee, H.J. Breathing Air in Air: In What Ways Might Extant Amphibious Fish Biology Relate to Prevailing Concepts about Early Tetrapods, the Evolution of Vertebrate Air Breathing, and the Vertebrate Land Transition? Physiol. Biochem. Zool. 2004, 77, 720–731. [Google Scholar] [CrossRef]
- Amemiya, C.T.; Alfoeldi, J.; Lee, A.P.; Fan, S.; Philippe, H.; Maccallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The African coelacanth genome provides insights into tetrapod evolution. Nat. Cell Biol. 2013, 496, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, M.; Noguchi, H.; Nishihara, H.; Toyoda, A.; Suzuki, Y.; Kajitani, R.; Suzuki, H.; Okuno, M.; Aibara, M.; Ngatunga, B.P.; et al. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013, 23, 1740–1748. [Google Scholar] [CrossRef] [Green Version]
- You, X.; Sun, M.; Li, J.; Bian, C.; Chen, J.; Yi, Y.; Yu, H.; Shi, Q. Mudskippers and Their Genetic Adaptations to an Amphibious Lifestyle. Animals 2018, 8, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [Green Version]
- Qiu, H.T.; Fernandes, J.M.; Hong, W.S.; Wu, H.X.; Zhang, Y.T.; Huang, S.; Liu, D.T.; Yu, H.; Wang, Q.; You, X.X. Paralogues from the expanded Tlr11 gene family in mudskipper (Boleophthalmus pectinirostris) are under positive selection and re-spond differently to LPS/Poly (I: C) challenge. Front. Immunol. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorente-Martínez, H.; Agorreta, A.; Torres-Sánchez, M.; Mauro, D.S. Evidence of positive selection suggests possible role of aquaporins in the water-to-land transition of mudskippers. Org. Divers. Evol. 2018, 18, 499–514. [Google Scholar] [CrossRef]
- Perutz, M.F. Species adaptation in a protein molecule. Mol. Biol. Evol. 1983, 1, 1–28. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an Improved Branch-Site Likelihood Method for Detecting Positive Selection at the Molecular Level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lespinet, O.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. The Role of Lineage-Specific Gene Family Expansion in the Evolution of Eukaryotes. Genome Res. 2002, 12, 1048–1059. [Google Scholar] [CrossRef] [Green Version]
- Sabbagh, A.; Marin, J.; Veyssière, C.; Lecompte, E.; Boukouvala, S.; Poloni, E.S.; Darlu, P.; Crouau-Roy, B. Rapid birth-and-death evolution of the xenobiotic metabolizing NAT gene family in vertebrates with evidence of adaptive selection. BMC Evol. Biol. 2013, 13, 62. [Google Scholar] [CrossRef] [Green Version]
- De Bie, T.; Cristianini, N.; DeMuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.V.; Thomas, G.W.; Lugo-Martinez, J.; Hahn, M.W. Estimating Gene Gain and Loss Rates in the Presence of Error in Genome Assembly and Annotation Using CAFE 3. Mol. Biol. Evol. 2013, 30, 1987–1997. [Google Scholar] [CrossRef]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löytynoja, A.; Goldman, N. Phylogeny-Aware Gap Placement Prevents Errors in Sequence Alignment and Evolutionary Analysis. Science 2008, 320, 1632–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug into decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes Empirical Bayes Inference of Amino Acid Sites under Positive Selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences: Fig. 1. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Van Dongen, S. A Cluster Algorithm For Graphs. Information Systems [INS]; CWI: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Janis, C.M.; Farmer, C. Proposed habitats of early tetrapods: Gills, kidneys, and the water-land transition. Zool. J. Linn. Soc. 1999, 126, 117–126. [Google Scholar] [CrossRef]
- Romero, P.E.; Weigand, A.M.; Pfenninger, M. Positive selection on panpulmonate mitogenomes provide new clues on adaptations to terrestrial life. BMC Evol. Biol. 2016, 16, 164. [Google Scholar] [CrossRef] [Green Version]
- Inohara, N.; Ogura, Y.; Chen, F.F.; Muto, A.; Nuñez, G. Human Nod1 confers responsiveness to bacterial lipopolysac-charides. J. Biol. Chem. 2001, 276, 2551–2554. [Google Scholar] [CrossRef] [Green Version]
- Bi, D.; Wang, Y.; Gao, Y.; Li, X.; Chu, Q.; Cui, J.; Xu, T. Recognition of Lipopolysaccharide and Activation of NF-κB by Cytosolic Sensor NOD1 in Teleost Fish. Front. Immunol. 2018, 9, 1413. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Aravind, L. The NACHT family–a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 2000, 25, 223–224. [Google Scholar] [CrossRef]
- Duan, B.; Davis, R.; Sadat, E.L.; Collins, J.; Sternweis, P.C.; Yuan, D.; Jiang, L.I. Distinct Roles of Adenylyl Cyclase VII in Regulating the Immune Responses in Mice. J. Immunol. 2010, 185, 335–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolanczyk, M.; Pech, M.; Zemojtel, T.; Yamamoto, H.; Mikula, I.; Calvaruso, M.-A.; Brand, M.V.D.; Richter, R.; Fischer, B.; Ritz, A.; et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell 2011, 22, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidler, J.; Al-Furoukh, N.; Kukat, C.; Salwig, I.; Ingelmann, M.-E.; Seibel, P.; Krueger, M.; Holtz, J.; Wittig, I.; Braun, T.; et al. Nitric Oxide-associated Protein 1 (NOA1) Is Necessary for Oxygen-dependent Regulation of Mitochondrial Respiratory Complexes. J. Biol. Chem. 2011, 286, 32086–32093. [Google Scholar] [CrossRef] [Green Version]
- Miner, J.H. Renal basement membrane components. Kidney Int. 1999, 56, 2016–2024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korstanje, R.; Deutsch, K.; Bolanos-Palmieri, P.; Hanke, N.; Schroder, P.; Staggs, L.; Bräsen, J.H.; Roberts, I.S.; Sheehan, S.; Savage, H.; et al. Loss of Kynurenine 3-Mono-oxygenase Causes Proteinuria. J. Am. Soc. Nephrol. 2016, 27, 3271–3277. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Sinha, R.P.; Häder, D.-P. UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 2002, 1, 225–236. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Rezabkova, L.; Wu, Z.; Wistow, G.; Schuck, P. Solution properties of γ-crystallins: Hydration of fish and mammal γ-crystallins. Protein Sci. 2014, 23, 88–99. [Google Scholar] [CrossRef] [Green Version]
- Hirano, M.; Das, S.; Guo, P.; Cooper, M.D. The Evolution of Adaptive Immunity in Vertebrates. Adv. Immunol. 2011, 109, 125–157. [Google Scholar] [CrossRef]
- Zhang, Y.-A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.J.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.L.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef]
- Mukherjee, T.; Hovingh, E.S.; Foerster, E.G.; Abdel-Nour, M.; Philpott, D.J.; Girardin, S.E. NOD1 and NOD2 in inflam-mation, immunity and disease. Arch. Biochem. Biophys. 2019, 670, 69–81. [Google Scholar] [CrossRef]
- Pashenkov, M.V.; Murugina, N.E.; Budikhina, A.S.; Pinegin, B.V. Synergistic interactions between NOD receptors and TLRs: Mechanisms and clinical implications. J. Leukoc. Biol. 2019, 105, 669–680. [Google Scholar] [CrossRef]
- Zhao, H.; Brown, P.H.; Magone, M.T.; Schuck, P. The Molecular Refractive Function of Lens γ-Crystallins. J. Mol. Biol. 2011, 411, 680–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slingsby, C.; Wistow, G.J. Functions of crystallins in and out of lens: Roles in elongated and post-mitotic cells. Prog. Biophys. Mol. Biol. 2014, 115, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakenhoff, R.H.; Aarts, H.J.; Reek, F.H.; Lubsen, N.H.; Schoenmakers, J.G. Human γ-crystallin genes: A gene family on its way to extinction. J. Mol. Biol. 1990, 216, 519–532. [Google Scholar] [CrossRef]
- Simpanya, M.F.; Wistow, G.; Gao, J.; David, L.L.; Giblin, F.J.; Mitton, K.P. Expressed sequence tag analysis of guinea pig (Cavia porcellus) eye tissues for NEIBank. Mol. Vis. 2008, 14, 2413–2427. [Google Scholar] [PubMed]
- Tomarev, S.; Segovia, L.; Slingsby, C.; Vihtelic, T. gammaN-crystallin and the evolution of the betagamma-crystallin su-per-family in vertebrates. FEBS J. 2005, 272, 22762291. [Google Scholar]
- Wistow, G. Lens crystallins: Gene recruitment and evolutionary dynamism. Trends Biochem. Sci. 1993, 18, 301–306. [Google Scholar] [CrossRef]
- Schwab, I.R. Janus on the mudflats. Br. J. Ophthalmol. 2003, 87, 13. [Google Scholar] [CrossRef] [Green Version]






| Scientific Name | Common Name | Taxon ID | Classification | Land Adaptation | Ensembl Assembly |
|---|---|---|---|---|---|
| Periophthalmus magnuspinnatus | Giant-fin mudskipper | 409849 | infraclass Teleostei | O | PM.fa |
| Homo sapiens | Human | 9606 | superclass Tetrapoda | O | GRCh38.p13 |
| Mus musculus | Mouse | 10090 | superclass Tetrapoda | O | GRCm38.p6 |
| Bos taurus | Cattle | 9913 | superclass Tetrapoda | O | ARS-UCD1.2 |
| Gallus gallus | Chicken | 9031 | superclass Tetrapoda | O | GRCg6a |
| Latimeria chalumnae | Coelacanth | 7897 | Non-tetrapod Sarcopterygii | X | LatCha1 |
| Danio rerio | Zebrafish | 7955 | infraclass Teleostei | X | GRCz11 |
| Astatotilapia calliptera | Eastern happy | 8154 | infraclass Teleostei | X | fAstCal1.2 |
| Amphiprion percula | Orange clownfish | 161767 | infraclass Teleostei | X | Nemo_v1 |
| Oryzias latipes | Japanese medaka | 8090 | infraclass Teleostei | X | ASM223471v1 |
| Parambassis ranga | Indian glassy fish | 210632 | infraclass Teleostei | X | fParRan2.1 |
| Poecilia formosa | Amazon molly | 48698 | infraclass Teleostei | X | Poecilia_formosa-5.1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, C.; Yoo, D.; Kim, H. Genetic Adaptations in Mudskipper and Tetrapod Give Insights into Their Convergent Water-to-Land Transition. Animals 2021, 11, 584. https://0-doi-org.brum.beds.ac.uk/10.3390/ani11020584
Kim J, Lee C, Yoo D, Kim H. Genetic Adaptations in Mudskipper and Tetrapod Give Insights into Their Convergent Water-to-Land Transition. Animals. 2021; 11(2):584. https://0-doi-org.brum.beds.ac.uk/10.3390/ani11020584
Chicago/Turabian StyleKim, Juwan, Chul Lee, DongAhn Yoo, and Heebal Kim. 2021. "Genetic Adaptations in Mudskipper and Tetrapod Give Insights into Their Convergent Water-to-Land Transition" Animals 11, no. 2: 584. https://0-doi-org.brum.beds.ac.uk/10.3390/ani11020584