Duplication, Loss, and Evolutionary Features of Specific UDP-Glucuronosyltransferase Genes in Carnivora (Mammalia, Laurasiatheria)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. UGT Phylogenetic Analysis and UGT Gene Counts
2.2. Synteny Analysis of UGT Genes
3. Results
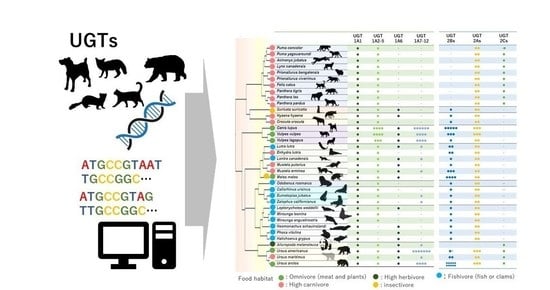
3.1. UGT Synteny Analysis and Gene Counts
3.1.1. UGT1A Coding Loci and Isoform Number in Mammals
3.1.2. UGT2A/B Coding Loci and Isoforms Number in Mammals
3.2. Phylogenetic Analysis of UGTs
3.2.1. Phylogenetic Analysis and Sequence Comparison of UGT1As in Carnivora
3.2.2. Phylogenetic Analysis of UGT2Bs in Carnivora
3.3. Sequence Comparison of UGT1As and 2Bs in Carnivora
4. Discussion
4.1. Relations between Diet and UGT2Bs Expansion
4.2. UGT1A Evolution and Adaptation to Species-Specific Diets
4.3. UGT Duplication/Loss and Relation to Functional Glucuronidation
4.4. Limitation of This Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andrew Williams, J.; Ring, B.J.; Cantrell, V.E.; Campanale, K.; Jones, D.R.; Hall, S.D.; Wrighton, S.A. Differential Modulation of UDP-Glucuronosyltransferase 1A1 (UGT1A1)-Catalyzed Estradiol-3-Glucuronidation by the Addition of UGT1A1 Substrates and Other Compounds to Human Liver Microsomes. Drug Metab. Dispos. 2002, 30, 1266–1273. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, J.R.; Kondapalli, R.; Chowdhury, N.R. Gunn Rat: A Model for Inherited Deficiency of Bilirubin Glucuronidation. Adv. Vet. Sci. Comp. Med. 1993, 37, 149–173. [Google Scholar]
- Lépine, J.; Bernard, O.; Plante, M.; Têtu, B.; Pelletier, G.; Labrie, F.; Bélanger, A.; Guillemette, C. Specificity and Regioselectivity of the Conjugation of Estradiol, Estrone, and Their Catecholestrogen and Methoxyestrogen Metabolites by Human Uridine Diphospho-Glucuronosyltransferases Expressed in Endometrium. J. Clin. Endocrinol. Metab. 2004, 89, 5222–5232. [Google Scholar] [CrossRef]
- Bock, K.W. Roles of Human UDP-Glucuronosyltransferases in Clearance and Homeostasis of Endogenous Substrates, and Functional Implications. Biochem. Pharmacol. 2015, 96, 77–82. [Google Scholar] [CrossRef]
- Tampal, N.; Lehmler, H.J.; Espandiari, P.; Malmberg, T.; Robertson, L.W. Glucuronidation of Hydroxylated Polychlorinated Biphenyls (PCBs). Chem. Res. Toxicol. 2002, 15, 1259–1266. [Google Scholar] [CrossRef]
- Daidoji, T.; Gozu, K.; Iwano, H.; Inoue, H.; Yokota, H. UDP-Glucuronosyltransferase Isoforms Catalyzing Glucuronidation of Hydroxy-Polychlorinated Biphenyls in Rat. Drug Metab. Dispos. 2005, 33, 1466–1476. [Google Scholar] [CrossRef] [Green Version]
- Kasai, N.; Sakaki, T.; Shinkyo, R.; Ikushiro, S.I.; Iyanagi, T.; Kamao, M.; Okano, T.; Ohta, M.; Inouye, K. Sequential Metabolism of 2,3,7-Trichlorodibenzo-p-Dioxin (2,3,7-TriCDD) by Cytochrome P450 and UDP-Glucuronosyltransferase in Human Liver Microsomes. Drug Metab. Dispos. 2004, 32, 870–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-Glucuronosyltransferases: Their Role in Drug Metabolism and Detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, C.; Lévesque, É.; Rouleau, M. Pharmacogenomics of Human Uridine Diphospho-Glucuronosyltransferases and Clinical Implications. Clin. Pharmacol. Ther. 2014, 96, 324–339. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, P.I.; Bock, K.W.; Burchell, B.; Guillemette, C.; Ikushiro, S.; Iyanagi, T.; Miners, J.O.; Owens, I.S.; Nebert, D.W. Nomenclature Update for the Mammalian UDP Glycosyltransferase (UGT) Gene Superfamily. Pharmacogenet. Genomics 2005, 15, 677–685. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Bélanger, A.; Fournel-Gigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP Glycosyltransferase Gene Superfamily: Recommended Nomenclature Update Based on Evolutionary Divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef]
- Li, C.; Wu, Q. Adaptive Evolution of Multiple-Variable Exons and Structural Diversity of Drug-Metabolizing Enzymes. BMC Evol. Biol. 2007, 7, 69. [Google Scholar] [CrossRef] [Green Version]
- Bock, K.W. Vertebrate UDP-Glucuronosyltransferases: Functional and Evolutionary Aspects. Biochem. Pharmacol. 2003, 66, 691–696. [Google Scholar] [CrossRef]
- Hassanin, A.; Veron, G.; Ropiquet, A.; van Vuuren, B.J.; Lécu, A.; Goodman, S.M.; Haider, J.; Nguyen, T.T. Evolutionary History of Carnivora (Mammalia, Laurasiatheria) Inferred from Mitochondrial Genomes. PLoS ONE 2021, 16, e0240770. [Google Scholar] [CrossRef]
- Werdelin, L.; Dehghani, R. Carnivora. In Paleontology and Geology of Laetoli: Human Evolution in Context; Vertebrate Paleobiology and Paleoanthropology Series; Springer: Cham, Switzerland, 2011; pp. 189–232. [Google Scholar] [CrossRef]
- Shrestha, B.; Reed, J.M.; Starks, P.T.; Kaufman, G.E.; Goldstone, J.V.; Roelke, M.E.; O’Brien, S.J.; Koepfli, K.P.; Frank, L.G.; Court, M.H. Evolution of a Major Drug Metabolizing Enzyme Defect in the Domestic Cat and Other Felidae: Phylogenetic Timing and the Role of Hypercarnivory. PLoS ONE 2011, 6, 221–237. [Google Scholar] [CrossRef] [Green Version]
- Kakehi, M.; Ikenaka, Y.; Nakayama, S.M.M.; Kawai, Y.K.; Watanabe, K.P.; Mizukawa, H.; Nomiyama, K.; Tanabe, S.; Ishizuka, M. Uridine Diphosphate-Glucuronosyltransferase (UGT) Xenobiotic Metabolizing Activity and Genetic Evolution in Pinniped Species. Toxicol. Sci. 2015, 147, 360–369. [Google Scholar] [CrossRef] [Green Version]
- Kondo, T.; Ikenaka, Y.; Nakayama, S.M.M.; Kawai, Y.K.; Mizukawa, H.; Mitani, Y.; Nomiyama, K.; Tanabe, S.; Ishizuka, M. Uridine Diphosphate-Glucuronosyltransferase (UGT) 2B Subfamily Interspecies Differences in Carnivores. Toxicol. Sci. 2017, 158, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Zhang, Y.; Zhang, P.; Liu, C.; Wang, J.; Gao, H.; Rus Hoelzel, A.; Seim, I.; Lv, M.; Lin, M.; et al. Comparative Genomics Provides Insights into the Aquatic Adaptations of Mammals. Proc. Natl. Acad. Sci. USA 2021, 118, e2106080118. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A Dynamic Web Platform for Genome Visualization and Analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef]
- Court, M.H.; Greenblatt, D.J. Molecular Genetic Basis for Deficient Acetaminophen Glucuronidation by Cats: UGT1A6 Is a Pseudogene, and Evidence for Reduced Diversity of Expressed Hepatic UGT1A Isoforms. Pharmacogenetics 2000, 10, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Blair Hedges, S. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W. The UDP-Glycosyltransferase (UGT) Superfamily Expressed in Humans, Insects and Plants: Animal-Plant Arms-Race and Co-Evolution. Biochem. Pharmacol. 2016, 99, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.J.; Nebert, D.W. Evolution of the P450 Gene Superfamily: Animal-Plant “Warfare”, Molecular Drive and Human Genetic Differences in Drug Oxidation. Trends Genet. 1990, 6, 182–186. [Google Scholar] [CrossRef]
- Wöll, S.; Kim, S.H.; Greten, H.J.; Efferth, T. Animal Plant Warfare and Secondary Metabolite Evolution. Nat. Prod. Bioprospect. 2013, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Kawai, Y.K.; Shinya, S.; Ikenaka, Y.; Saengtienchai, A.; Kondo, T.; Darwish, W.S.; Nakayama, S.M.M.; Mizukawa, H.; Ishizuka, M. Characterization of Function and Genetic Feature of UDP-Glucuronosyltransferase in Avian Species. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 217, 5–14. [Google Scholar] [CrossRef]
- Greenhalgh, R.; Holding, M.L.; Orr, T.J.; Henderson, J.B.; Parchman, T.L.; Matocq, M.D.; Shapiro, M.D.; Dearing, M.D. Trio-Binned Genomes of the Woodrats Neotoma Bryanti and Neotoma Lepida Reveal Novel Gene Islands and Rapid Copy Number Evolution of Xenobiotic Metabolizing Genes. Mol. Ecol. Resour. 2022, 22, 2713–2731. [Google Scholar] [CrossRef]
- Xing, X.; Ai, C.; Wang, T.; Li, Y.; Liu, H.; Hu, P.; Wang, G.; Liu, H.; Wang, H.; Zhang, R.; et al. The First High-Quality Reference Genome of Sika Deer Provides Insights for High-Tannin Adaptation. Genom. Proteom. Bioinform. 2022, in press. [Google Scholar] [CrossRef]
- Kawai, Y.K.; Ikenaka, Y.; Ishizuka, M.; Kubota, A. The Evolution of UDP-Glycosyl/Glucuronosyltransferase 1E (UGT1E) Genes in Bird Lineages Is Linked to Feeding Habits but UGT2 Genes Is Not. PLoS ONE 2018, 13, e0205266. [Google Scholar] [CrossRef]
- Kawai, Y.K.; Sano, K.; Ikenaka, Y.; Nakayama, S.M.M.; Kondo, M.; Kubota, A.; Ishizuka, M. Evolutionary History of Mammalian UDP-Glucuronosyltransferase (UGT)1 and UGT2 Families: The Emergence of UGT2B Subfamily in Eutherians after the Diversification of Flowering Plants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dell’Arte, G.L.; Laaksonen, T.; Norrdahl, K.; Korpimäki, E. Variation in the Diet Composition of a Generalist Predator, the Red Fox, in Relation to Season and Density of Main Prey. Acta Oecol. 2007, 31, 276–281. [Google Scholar] [CrossRef]
- Baltrūnaitė, L.; Baltrûnaitë, L. Diet Composition of the Red Fox (Vulpes vulpes L.), Pine Marten (Martes martes L.) and Raccoon Dog (Nyctereutes procyonoides Gray) in Clay Plain Landscape, Lithuania. Acta Zool. Litu. 2012, 12, 362–368. [Google Scholar] [CrossRef]
- Merkle, J.A.; Polfus, J.L.; Derbridge, J.J.; Heinemeyer, K.S. Dietary Niche Partitioning among Black Bears, Grizzly Bears, and Wolves in a Multiprey Ecosystem. Can. J. Zool. 2017, 95, 663–671. [Google Scholar] [CrossRef]
- Costello, C.M.; Cain, S.L.; Pils, S.; Frattaroli, L.; Haroldson, M.A.; van Manen, F.T. Diet and Macronutrient Optimization in Wild Ursids: A Comparison of Grizzly Bears with Sympatric and Allopatric Black Bears. PLoS ONE 2016, 11, e0153702. [Google Scholar] [CrossRef] [PubMed]
- Hilderbrand, G.V.; Farley, S.D.; Robbins, C.T.; Hanley, T.A.; Titus, K.; Servheen, C. Use of Stable Isotopes to Determine Diets of Living and Extinct Bears. Can. J. Zool. 1996, 74, 2080–2088. [Google Scholar] [CrossRef]
- Jiangzuo, Q.; Flynn, J.J. The Earliest Ursine Bear Demonstrates the Origin of Plant-Dominated Omnivory in Carnivora. iScience 2020, 23, 101235. [Google Scholar] [CrossRef]
- Lazarus, M.; Sekovanić, A.; Orct, T.; Reljić, S.; Kusak, J.; Jurasović, J.; Huber, Đ. Apex Predatory Mammals as Bioindicator Species in Environmental Monitoring of Elements in Dinaric Alps (Croatia). Environ. Sci. Pollut. Res. 2017, 24, 23977–23991. [Google Scholar] [CrossRef]
- Gagliano, J.; Anselmo-Moreira, F.; Sala-Carvalho, W.R.; Furlan, C.M. What Is Known about the Medicinal Potential of Bamboo? Adv. Tradit. Med. 2022, 22, 467–495. [Google Scholar] [CrossRef]
- Johnson, R.N.; O’Meally, D.; Chen, Z.; Etherington, G.J.; Ho, S.Y.W.; Nash, W.J.; Grueber, C.E.; Cheng, Y.; Whittington, C.M.; Dennison, S.; et al. Adaptation and Conservation Insights from the Koala Genome. Nat. Genet. 2018, 50, 1102–1111. [Google Scholar] [CrossRef]
- Kitanovic, S.; Marks-Fife, C.A.; Parkes, Q.A.; Wilderman, P.R.; Halpert, J.R.; Dearing, M.D. Cytochrome P450 2B Diversity in a Dietary Specialist-the Red Tree Vole (Arborimus longicaudus). J. Mammal. 2018, 99, 578–585. [Google Scholar] [CrossRef]
- Kitanovic, S.; Orr, T.J.; Spalink, D.; Cocke, G.B.; Schramm, K.; Wilderman, P.R.; Halpert, J.R.; Dearing, M.D. Role of Cytochrome P450 2B Sequence Variation and Gene Copy Number in Facilitating Dietary Specialization in Mammalian Herbivores. Mol. Ecol. 2018, 27, 723–736. [Google Scholar] [CrossRef]
- Huang, G.; Wang, X.; Hu, Y.; Wu, Q.; Nie, Y.; Dong, J.; Ding, Y.; Yan, L.; Wei, F. Diet Drives Convergent Evolution of Gut Microbiomes in Bamboo-Eating Species. Sci. China Life Sci. 2020, 64, 88–95. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, Q.; Dai, J.; Zhang, S.; Wei, F. Evidence of Cellulose Metabolism by the Giant Panda Gut Microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 17714–17719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Wang, L.; Li, J.; Hou, R.; Wang, M.; Wang, Z.; Qu, Q.; Zhou, W.; Nie, Y.; Hu, Y.; et al. Seasonal Shift of the Gut Microbiome Synchronizes Host Peripheral Circadian Rhythm for Physiological Adaptation to a Low-Fat Diet in the Giant Panda. Cell Rep. 2022, 38, 110203. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, E.; Ratnakumar, A.; Arendt, M.L.; Maqbool, K.; Webster, M.T.; Perloski, M.; Liberg, O.; Arnemo, J.M.; Hedhammar, Å.; Lindblad-Toh, K. The Genomic Signature of Dog Domestication Reveals Adaptation to a Starch-Rich Diet. Nature 2013, 495, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Doncaster, C.P.; Dickman, C.R.; Macdonald, D.W. Feeding Ecology of Red Foxes (Vulpes vulpes) in the City of Oxford, England. J. Mammal. 1990, 71, 188–194. [Google Scholar] [CrossRef]
- Soe, E.; Davison, J.; Süld, K.; Valdmann, H.; Laurimaa, L.; Saarma, U. Europe-Wide Biogeographical Patterns in the Diet of an Ecologically and Epidemiologically Important Mesopredator, the Red Fox Vulpes vulpes: A Quantitative Review. Mamm. Rev. 2017, 47, 198–211. [Google Scholar] [CrossRef]
- Elmhagen, B.; Tannerfeldt, M.; Verucci, P.; Angerbjörn, A. The Arctic Fox (Alopex lagopus): An Opportunistic Specialist. J. Zool. 2000, 251, 139–149. [Google Scholar] [CrossRef]
- Balestrieri, A.; Remonti, L.; Saino, N.; Raubenheimer, D. The ‘Omnivorous Badger Dilemma’: Towards an Integration of Nutrition with the Dietary Niche in Wild Mammals. Mamm. Rev. 2019, 49, 324–339. [Google Scholar] [CrossRef]
- Westbury, M.V.; Le Duc, D.; Duchêne, D.A.; Krishnan, A.; Prost, S.; Rutschmann, S.; Grau, J.H.; Dalén, L.; Weyrich, A.; Norén, K.; et al. Ecological Specialization and Evolutionary Reticulation in Extant Hyaenidae. Mol. Biol. Evol. 2021, 38, 3884–3897. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.B.; Klaassen, C.D. Tissue- and Gender-Specific MRNA Expression of UDP-Glucuronosyltransferases (UGTs) in Mice. Drug Metab. Dispos. 2007, 35, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sneitz, N.; Court, M.H.; Zhang, X.; Laajanen, K.; Yee, K.K.; Dalton, P.; Ding, X.; Finel, M. Human UDP-Glucuronosyltransferase UGT2A2: CDNA Construction, Expression, and Functional Characterization in Comparison with UGT2A1 and UGT2A3. Pharmacogenet. Genomics 2009, 19, 923–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soars, M.G.; Riley, R.J.; Findlay, K.A.; Coffey, M.J.; Burchell, B. Evidence for Significant Differences in Microsomal Drug Glucuronidation by Canine and Human Liver and Kidney. Drug Metab. Dispos. 2001, 29, 121–126. [Google Scholar] [PubMed]
- Court, M.H. Feline Drug Metabolism and Disposition: Pharmacokinetic Evidence for Species Differences and Molecular Mechanisms. Vet. Clin. N. Am. Small Anim. Pract. 2013, 43, 1039–1054. [Google Scholar] [CrossRef] [Green Version]
- Redmon, J.M.; Shrestha, B.; Cerundolo, R.; Court, M.H. Soy Isoflavone Metabolism in Cats Compared with Other Species: Urinary Metabolite Concentrations and Glucuronidation by Liver Microsomes. Xenobiotica 2016, 46, 406–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Beusekom, C.D.; Fink-Gremmels, J.; Schrickx, J.A. Comparing the Glucuronidation Capacity of the Feline Liver with Substrate-Specific Glucuronidation in Dogs. J. Vet. Pharmacol. Ther. 2014, 37, 18–24. [Google Scholar] [CrossRef]
- Owens, I.S.; Basu, N.K.; Banerjee, R. UDP-Glucuronosyltransferases: Gene Structures of UGT1 and UGT2 Families. Methods Enzymol. 2005, 400, 1–22. [Google Scholar] [CrossRef]
- Guillemette, C.; Lévesque, E.; Harvey, M.; Bellemare, J.; Menard, V. UGT Genomic Diversity: Beyond Gene Duplication. Drug Metab. Rev. 2010, 42, 24–44. [Google Scholar] [CrossRef]
- Lu, H.; Gunewardena, S.; Cui, J.Y.; Yoo, B.; Zhong, X.B.; Klaassen, C.D. RNA-Sequencing Quantification of Hepatic Ontogeny and Tissue Distribution of MRNAs of Phase II Enzymes in Mice. Drug Metab. Dispos. 2013, 41, 844–857. [Google Scholar] [CrossRef] [Green Version]
- Uno, Y.; Takahira, R.; Murayama, N.; Onozeki, S.; Kawamura, S.; Uehara, S.; Ikenaka, Y.; Ishizuka, M.; Ikushiro, S.; Yamazaki, H. Functional and Molecular Characterization of UDP-Glucuronosyltransferase 2 Family in Cynomolgus Macaques. Biochem. Pharmacol. 2019, 163, 335–344. [Google Scholar] [CrossRef]
- Sakai, C.; Iwano, S.; Shimizu, M.; Onodera, J.; Uchida, M.; Sakurada, E.; Yamazaki, Y.; Asaoka, Y.; Imura, N.; Uno, Y.; et al. Analysis of Gene Expression for Microminipig Liver Transcriptomes Using Parallel Long-Read Technology and Short-Read Sequencing. Biopharm. Drug Dispos. 2016, 37, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Thibaud-Nissen, F.; Souvorov, A.; Murphy, T.; DiCuccio, M.; Kitts, P. Eukaryotic Genome Annotation Pipeline. In The NCBI Handbook, 2nd ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2013. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, M.; Ikenaka, Y.; Nakayama, S.M.M.; Kawai, Y.K.; Ishizuka, M. Duplication, Loss, and Evolutionary Features of Specific UDP-Glucuronosyltransferase Genes in Carnivora (Mammalia, Laurasiatheria). Animals 2022, 12, 2954. https://0-doi-org.brum.beds.ac.uk/10.3390/ani12212954
Kondo M, Ikenaka Y, Nakayama SMM, Kawai YK, Ishizuka M. Duplication, Loss, and Evolutionary Features of Specific UDP-Glucuronosyltransferase Genes in Carnivora (Mammalia, Laurasiatheria). Animals. 2022; 12(21):2954. https://0-doi-org.brum.beds.ac.uk/10.3390/ani12212954
Chicago/Turabian StyleKondo, Mitsuki, Yoshinori Ikenaka, Shouta M. M. Nakayama, Yusuke K. Kawai, and Mayumi Ishizuka. 2022. "Duplication, Loss, and Evolutionary Features of Specific UDP-Glucuronosyltransferase Genes in Carnivora (Mammalia, Laurasiatheria)" Animals 12, no. 21: 2954. https://0-doi-org.brum.beds.ac.uk/10.3390/ani12212954