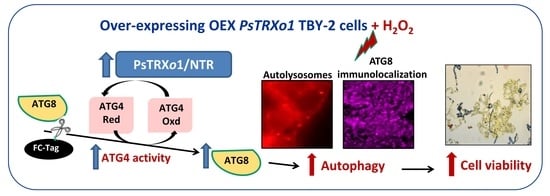
Autophagy Is Involved in the Viability of Overexpressing Thioredoxin o1 Tobacco BY-2 Cells under Oxidative Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transformation of Tobacco BY-2 Cells
2.2. Cell Viability
2.3. H2O2 Treatment
2.4. Incubation in the Presence of Autophagy Inhibitors
2.5. Protein Extracts
2.6. Quantitative Real-Time PCR
2.7. SDS–PAGE and Western Blot of Autophagy Markers
2.8. ATG4 Cleavage Assay
2.9. Visualization of Autolysosomes
2.10. Immunolocalization of ATG8
2.11. Protein Dot-Blot
2.12. Co-Immunoprecipitation PsTRXo1/His-HsATG4
2.13. Statistical Analysis
3. Results
3.1. Overexpressing PsTRXo1 Cells Present High Viability and Elevated Levels of Autophagy Markers
3.2. Autophagy Is Related to Cell Viability
3.3. Overexpressing Pstrxo1 Cells Present Lytic Vesicles 24 h and 48 h after Oxidative Treatment
3.4. Overexpressing Pstrxo1 Cells Accumulate ATG8 Protein Detected by Immunofluorescence 24 h after Oxidative Treatment
3.5. Autophagy Is Evident in OEX Cells and Not in Control Cells 14 h after Oxidative Treatment
3.6. ATG4 and TRXo1 Interact In Vitro
3.7. ATG4 Activity Is Redox-Regulated
3.8. ATG4 Activity Is Redox-Regulated by Thioredoxin o1 In Vitro
4. Discussion
4.1. Autophagy, Oxidative Stress and TRX
4.2. Autophagy, Cell Survival and TRX
4.3. TRX-Dependent Redox Regulation of Autophagy Target Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dodson, M.; Darley-Usmar, V.; Zhang, J. Cellular metabolic and autophagic pathways: Traffic control by redox signaling. Free Rad. Biol. Med. 2013, 63, 207–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Klionsky, D. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010, 12, 814–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marion, J.; Le-Bars, R.; Besse, L.; Batoko, H.; Satiat-Jeunemaitre, B. Multiscale and multimodal approaches to study autophagy in model plants. Cells 2018, 7, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, Y.; Moriyasu, Y. Autophagy is not a main contributor to the degradation of phospholipids in tobacco cells cultured under sucrose starvation conditions. Plant Cell Physiol. 2006, 47, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Takatsuka, C.; Inoue, Y.; Higuchi, T.; Hillmer, S.; Robinson, D.G.; Moriyasu, Y. Autophagy in tobacco BY-2 cells cultured under sucrose starvation sonditions: Isolation of the autolysosome and its caracterization. Plant Cell Physiol. 2011, 52, 2074–2087. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J. Teaching the basics of autophagy and mitophagy to redox biologists—Mechanisms and experimental approaches. Redox Biol. 2015, 4, 242–259. [Google Scholar] [CrossRef] [Green Version]
- Aroca, A.; Yruela, I.; Gotor, C.; Bassham, D.C. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023604118. [Google Scholar] [CrossRef]
- Zhang, J. Autophagy and mitophagy in cellular damage control. Redox Biol. 2013, 1, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Marshal, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 9, 173–208. [Google Scholar] [CrossRef]
- Üstün, S.; Hafrén, A.; Hofius, D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 2017, 40, 122–130. [Google Scholar] [CrossRef]
- Yu, Z.Q.; Ni, T.; Hong, B.; Wang, H.Y.; Jiang, F.J.; Zou, S.; Xie, Z. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 2012, 8, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Seo, E.; Woo, J.; Park, E.; Bertolani, S.J.; Siegel, J.B.; Choi, D.; Dinesh-Kumar, S.P. Comparative analysis of ubiquitin-like ATG8 and cysteine protease ATG4 autophagy genes in the plant lineage and cross-kingdom processing of ATG8 by ATG4. Autophagy 2016, 12, 2054–2068. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.M.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.h.; Peng, X.B.; Sun, M.X. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Sevilla, F.; Camejo, D.; Ortiz-Espín, A.; Calderón, A.; Lázaro, J.J.; Jiménez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef] [Green Version]
- Zannini, F.; Roret, T.; Przybyla-Toscano, J.; Dhalleine, T.; Rouhier, N.; Couturier, J. Mitochondrial Arabidopsis thaliana TRXo isoforms bind an iron–sulfur cluster and reduce NFU proteins in vitro. Antioxidants 2018, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Cejudo, F.J.; Ojeda, V.; Delgado-Requerey, V.; González, M.; Pérez-Ruiz, J.M. Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front. Plant Sci. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Martí, M.C.; Jiménez, A.; Sevilla, F. Thioredoxin network in plant mitochondria: Cysteine S-posttranslational modifications and stress conditions. Front. Plant Sci. 2020, 11, 571288. [Google Scholar] [CrossRef]
- Shao, F.-Y.; Du, Z.-Y.; Ma, D.-L.; Chen, W.-B.; Fu, W.-Y.; Ruan, B.-B.; Rui, W.; Zhang, Z.-X.; Wang, X.; Wong, N.S. B5, a thioredoxin reductase inhibitor, induces apoptosis in human cervical cancer cells by suppressing the thioredoxin system, disrupting mitochondrion-dependent pathways and triggering autophagy. Oncotarget 2015, 6, 30939–30956. [Google Scholar] [CrossRef] [Green Version]
- Nagakannan, P.; Iqbal, M.A.; Yeung, A.; Thliveris, J.A.; Rastegar, M.; Ghavami, S.; Eftekharpour, E. Perturbation of redox balance after thioredoxin reductase deficiency interrupts autophagy-lysosomal degradation pathway and enhances cell death in nutritionally stressed SH-SY5Y cells. Free Radic. Biol. Med. 2016, 101, 53–70. [Google Scholar] [CrossRef]
- Picazo, C.; Matallana, E.; Aranda, A. Yeast thioredoxin reductase Trr1p controls TORC1-regulated processes. Sci. Rep. 2018, 8, 16500. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.; Zaffagnini, M.; Marchand, C.H.; Crespo, J.L.; Lemaire, S.D. The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 2014, 10, 1953–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Pérez, M.; Lemaire, S.D.; Crespo, J.L. Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 2016, 172, 2219–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martí, M.C.; Olmos, E.; Calvete, J.J.; Diaz, I.; Barranco-Medina, S.; Whelan, J.; Jiménez, A. Mitochondrial and nuclear localization of a novel pea thioredoxin: Identification of its mitochondrial target proteins. Plant Physiol. 2009, 150, 646–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barranco-Medina, S.; Krell, T.; Finkemeier, I.; Sevilla, F.; Lázaro, J.J.; Dietz, K.J. Biochemical and molecular characterization of the mitochondrial peroxiredoxin PsPrxII F from Pisum sativum. Plant Physiol. Biochem. 2007, 45, 729–739. [Google Scholar] [CrossRef]
- Martí, M.C.; Florez-Sarasa, I.; Camejo, D.; Ribas-Carbó, M.; Lázaro, J.J.; Sevilla, F.; Jiménez, A. Response of the mitochondrial antioxidant redox system and respiration to salinity in pea plants. J. Exp. Bot. 2011, 2, 3863–3874. [Google Scholar] [CrossRef] [Green Version]
- Calderón, A.; Ortiz-Espín, A.; Iglesias-Fernández, R.; Carbonero, P.; Pallardó, F.; Sevilla, F.; Jiménez, A. Thioredoxin (Trxo1) interacts with proliferating cell nuclear antigen (PCNA) and its overexpression affects the growth of tobacco cell culture. Redox Biol. 2017, 11, 688–700. [Google Scholar] [CrossRef]
- Da Fonseca-Pereira, P.; Daloso, D.M.; Gago, J.; Magnum, F.; Silva, O.; Condori-Apfata, J.A.; Florez-Sarasa, I.; Araújo, W.L. The mitochondrial thioredoxin system contributes to the metabolic responses under drought episodes in Arabidopsis. Plant Cell Physiol. 2019, 60, 213–229. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Espín, A.; Locato, V.; Camejo, D.; Schiermeyer, A.; De Gara, L.; Sevilla, F.; Jiménez, A. Over-expression of Trxo1 increases the viability of tobacco BY-2 cells under H2O2 treatment. Ann. Bot. 2015, 116, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef]
- Bárány, I.; Berenguer, E.; Solís, M.T.; Pérez-Pérez, Y.; Santamaría, M.E.; Crespo, J.L.; Testillano, P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018, 69, 1387–1402. [Google Scholar] [CrossRef] [Green Version]
- Lenz, H.D.; Haller, E.; Melzer, E.; Gust, A.A.; Nürnberger, T. Autophagy controls plant basal immunity in a pathogenic lifestyle-dependent manner. Autophagy 2011, 7, 773–774. [Google Scholar] [CrossRef] [Green Version]
- Takatsuka, C.; Inoue, Y.; Matsuoka, K.; Moriyasu, Y. 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 2004, 45, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- López-Vidal, O.; Olmedilla, A.; Sandalio, L.M.; Sevilla, F.; Jiménez, A. Is autophagy involved in pepper fruit ripening? Cells 2020, 9, 106. [Google Scholar] [CrossRef] [Green Version]
- Voitsekhovskaja, O.V.; Schiermeyer, A.; Reumann, S. Plant peroxisomes are degraded by starvation-induced and constitutive autophagy in tobacco BY-2 suspension-cultured cells. Front. Plant Sci. 2014, 5, 629. [Google Scholar] [CrossRef] [Green Version]
- Álvarez, C.; García, I.; Moreno, I.; Pérez-Pérez, M.E.; Crespo, J.L.; Romero, L.C.; Gotor, C. Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 2012, 24, 4621–4634. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.H.; Yoshimoto, K.; Ohsumi, Y.; Jeon, J.S.; An, G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 2009, 27, 67–74. [Google Scholar] [CrossRef]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pérez, M.E.; Florencio, F.J.; Crespo, J.L. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010, 152, 1874–1888. [Google Scholar] [CrossRef] [Green Version]
- Calero-Muñoz, N.; Exposito-Rodriguez, M.; Collado-Arenal, A.M.; Rodríguez-Serrano, M.; Laureano-Marín, A.M.; Santamaría, M.E.; Sandalio, L.M. Cadmium induces reactive oxygen species-dependent pexophagy in Arabidopsis leaves. Plant Cell Environ. 2019, 42, 2696–2714. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An intracellular degradation pathway regulating plant survival and stress response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pengo, N.; Agrotis, A.; Prak, K.; Jones, J.; Ketteler, R. A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat. Commun. 2017, 8, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivaraj, S.M.; Vats, S.; Bhat, J.A.; Dhakte, P.; Goyal, V.; Khatri, P.; Deshmukh, R. Nitric oxide and hydrogen sulfide crosstalk during heavy metal stress in plants. Physiol. Plant. 2020, 168, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Laureano-Marín, A.M.; Aroca, A.; Pérez-Pérez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Gotor, C. Abscisic acid-triggered persulfidation of the Cys protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. Regulation of autophagy by ROS: Physiology and pathology. Trends Biochem. Sci. 2011, 36, 30–38. [Google Scholar] [CrossRef]
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11. [Google Scholar] [CrossRef]
- Yano, K.; Suzuki, T.; Moriyasu, Y. Constitutive autophagy in plant root cells. Autophagy 2007, 3, 360–362. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Gong, Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Contento, A.L.; Nguyen, P.Q.; Bassham, D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007, 143, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin Md., J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [Green Version]
- Couso, I.; Pérez-Pérez, M.E.; Martínez-Force, E.; Kim, H.-S.; He, Y.; Umen, J.G.; Crespo, J.L. Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J. Exp. Bot. 2018, 69, 1355–1367. [Google Scholar] [CrossRef]
- Bassham, D.C. Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593. [Google Scholar] [CrossRef]
- Goto-Yamada, S.; Oikawa, K.; Bizan, J.; Shigenobu, S.; Yamaguchi, K.; Mano, S.; Yamada, K. Sucrose starvation induces microautophagy in plant root cells. Front. Plant Sci. 2019, 10, 1604. [Google Scholar] [CrossRef] [Green Version]
- Dauphinee, A.N.; Denbigh, G.L.; Rollini, A.; Fraser, M.; Lacroix, C.R.; Gunawardena, A. The function of autophagy in lace plant programmed cell death. Front. Plant Sci. 2019, 10, 1198. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vicencio, J.M.; Keep, O.; Tasdemir, E.; Maiuri, M.C.; Kroemer, G. To die or not to die: That is the autophagic question. Curr. Mol. Med. 2008, 8, 78–91. [Google Scholar]
- Simonsen, A.; Cumming, R.C.; Brech, A.; Isakson, P.; Schubert, D.R.; Finley, K.D. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 2008, 4, 176–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radyuk, S.N.; Michalak, K.; Klichko, V.I.; Benes, J.; Rebrin, I.; Sohal, R.S.; Orr, W.C. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. Biochem. J. 2009, 419, 437–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radyuk, S.N.; Rebrin, I.; Klichko, V.I.; Sohal, B.H.; Michalak, K.; Benes, J.; Orr, W.C. Mitochondrial peroxiredoxins are critical for the maintenance of redox state and the survival of adult Drosophila. Free Radic. Biol. Med. 2010, 49, 1892–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, A.R.; Suttangkakul, A.; Vierstra, R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008, 178, 1339–1353. [Google Scholar] [CrossRef] [Green Version]
- Doherty, J.; Baehrecke, E.H. Life, death and autophagy. Nat. Cell Biol. 2018, 20, 1110–1117. [Google Scholar] [CrossRef]
- Zhou, J.; Yao, K.; Zhang, Y.; Chen, G.; Lai, K.; Yin, H.; Yu, Y. Thioredoxin binding protein-2 regulates autophagy of human lens epithelial cells under oxidative stress via inhibition of Akt phosphorylation. Oxid. Med. Cell Longev. 2016, 856431, 1–17. [Google Scholar] [CrossRef]
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef]
- Bian, M.; Fan, R.; Zhao, S.; Liu, W. Targeting the thioredoxin system as a strategy for cancer therapy. J. Med. Chem. 2019, 62, 7309–7321. [Google Scholar] [CrossRef]
- Woo, J.; Park, E.; Dinesh-Kumar, S.P. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc. Nat. Acad. Sci. USA 2014, 111, 863–868. [Google Scholar] [CrossRef] [Green Version]
- Cochemé, H.M.; Bjedov, I.; Grönke, S.; Menger, K.E.; James, A.M.; Castillo-Quan, J.I.; Partridge, L. Enhancing autophagy by redox regulation extends lifespan in Drosophila. BioRxiv 2019, 790378. [Google Scholar]
- Oka, S.; Hirata, T.; Suzuki, W.; Naito, D.; Chen, Y.; Chin, A.; Sadoshima, J. Thioredoxin-1 maintains mechanistic target of rapamycin (mTOR) function during oxidative stress in cardiomyocytes. J. Biol. Chem. 2017, 292, 8988–19000. [Google Scholar] [CrossRef] [Green Version]
- Frudd, K.; Burgoyne, T.; Burgoyne, J.R. Oxidation of Atg3 and Atg7 mediates inhibition of autophagy. Nat. Commun. 2018, 9, 95. [Google Scholar] [CrossRef] [Green Version]













Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Brasi-Velasco, S.; López-Vidal, O.; Martí, M.C.; Ortiz-Espín, A.; Sevilla, F.; Jiménez, A. Autophagy Is Involved in the Viability of Overexpressing Thioredoxin o1 Tobacco BY-2 Cells under Oxidative Conditions. Antioxidants 2021, 10, 1884. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10121884
De Brasi-Velasco S, López-Vidal O, Martí MC, Ortiz-Espín A, Sevilla F, Jiménez A. Autophagy Is Involved in the Viability of Overexpressing Thioredoxin o1 Tobacco BY-2 Cells under Oxidative Conditions. Antioxidants. 2021; 10(12):1884. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10121884
Chicago/Turabian StyleDe Brasi-Velasco, Sabrina, Omar López-Vidal, María Carmen Martí, Ana Ortiz-Espín, Francisca Sevilla, and Ana Jiménez. 2021. "Autophagy Is Involved in the Viability of Overexpressing Thioredoxin o1 Tobacco BY-2 Cells under Oxidative Conditions" Antioxidants 10, no. 12: 1884. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10121884