1. Introduction
Pathogenic bacteria employ a variety of mechanisms to overcome the host’s immune response and to spread infections. The most-studied and relevant mechanism is arguably the quorum-sensing system, which detects cell density through cell-to-cell communication mechanisms and consequently regulates the expression of specific genes responsible for immune evasion and virulence. Numerous processes are controlled by quorum-sensing in bacteria, such as sporulation, gene transfer, biofilm formation, and virulence factor secretion [
1].
Staphylococcus aureus (
S. aureus) is a Gram-positive bacterium normally found on human skin and mucous membranes. It is an opportunistic human pathogen that causes serious infections that can lead to acute and chronic illnesses and have life-threatening consequences [
2]. The accessory gene regulator (
agr) quorum-sensing system allows for the rapid adaptation of
S. aureus to environmental changes and thus promotes virulence and the development of pathogenesis [
3]. This two-component system induces the transcription of
rnaII and
rnaIII via the P2 and P3 promoters, respectively. The P2 promoter governs the expression of the
agr operon, which contains four genes:
agrA, agrB,
agrC, and
agrD. AgrA is the response regulator that binds to both P2 and P3 promoters and upregulates the transcription of
agr and
rnaIII, respectively. It is activated by AgrC, a transmembrane histidine kinase that is activated by autophosphorylation upon binding to the autoinducing peptide (AIP). AgrB is a transmembrane endopeptidase that cleaves the C-terminus of the AIP precursor (encoded by
agrD), introduces a thiolactone bond between the C-terminus and an internal cysteine of AIP, and exports active AIP into the extracellular space. Indeed, AgrC and AgrA are two components of a signal transduction pathway that is activated in the late log phase of bacterial growth when the concentration of extracellular AIP is high. Increased transcription from the P3 promoter by AgrA activates RNAIII expression (the
agr effector molecule), which itself regulates the expression of several genes responsible for virulence and immune evasion. As a result, the expression of cell surface components decreases and the transcription of virulence factors (such as hemolysins and TSS toxin-1) increases, which allows bacteria that have reached high densities to acquire more nutrients from the host and spread the infection further [
3]. In addition to AIP, the
agr system can also be activated in response to extracellular stimuli such as glucose concentration and pH, as well as transcriptional regulators SarA and SrrAB [
4].
The
agr quorum response was shown to be sensitive to oxidizing conditions, mediated by an intramolecular disulfide switch in the DNA-binding domain of AgrA [
5]. AgrA contains two structural domains that mediate its transcriptional activity: the response regulator and a DNA-binding (LytTR) domain [
6]. Cysteine (Cys) 199 was identified as crucial for oxidation sensing and the formation of a disulfide bond with Cys228 upon oxidative stress, which inhibits AgrA binding to DNA [
5]. It has been proposed that the formation of an intramolecular disulfide in AgrA during oxidizing conditions induces a conformational change in the DNA-binding domain, leading to a steric interference and the dissociation of AgrA from DNA.
Cysteine residues on proteins are well-known targets of various oxidative post-translational modifications (oxPTMs), including S-thiolation, nitrosation, acetylation, and persulfhydration, among others. A range of enzymes, receptors, and transcription factors are regulated by oxPTMs [
7]. Protein glutathionylation (the thiolation of cysteine residues by glutathione (GSH)) is the most studied form of S-thiolation and occurs in response to oxidative, nitrosative, or metabolic stress. It has a variety of functions, including protecting protein cysteines from irreversible overoxidation to the sulfonic forms [
8]. Protein glutathionylation was shown to modulate regulatory interactions, DNA binding, and transcriptional activities of many prokaryotic transcriptional regulators that are involved in bacterial redox regulation and the adaptation to stresses [
9,
10]. GSH is produced in all Gram-negative bacteria, but not in Gram-positive
Firmicutes (including
Bacillus and
Staphylococcus) and
Actinomycetes species that synthesize bacillithiol (BSH) and mycothiol, respectively [
8]. In
Bacillus subtilis, bacillithiolation was found to regulate the activity of the redox-sensing OhrR repressor [
11,
12], whereas mycothiolation of several key enzymes was also described in
Actinomycetes [
13].
Coenzyme A (CoA) is another major thiol produced in all living cells by enzymatic conjugation of ATP, pantothenate (vitamin B5), and cysteine [
14,
15]. CoA and CoA thioesters participate in diverse anabolic and catabolic pathways, including the citric acid cycle, fatty acid biosynthesis and oxidation, amino acid metabolism, and isoprenoid and peptidoglycan biosynthesis. The antioxidant function of CoA has been recently reported in both eukaryotic and prokaryotic cells [
16,
17]. The development of novel research tools and methodologies has allowed the identification and characterization of CoA modified proteins (CoAlation) in vitro and in cell-based and animal models [
14,
18,
19]. Protein CoAlation was found to be a widespread and reversible oxPTM [
20,
21]. To date, over 2100 CoAlated proteins have been identified by the developed mass spectrometry-based methodology in mammalian cells and bacteria exposed to oxidative or metabolic stress. CoAlation was found to modulate the activity and conformation of modified proteins, and protect key cysteine residues from overoxidation [
18,
19]. Bioinformatics analysis of CoAlated proteins in mammalian cells revealed that the majority (over 65%) are involved in metabolic processes. In contrast to mammalian cells, numerous transcription factors and regulators were found to be CoAlated in
S. aureus and
Bacillus megaterium (
B. megaterium) exposed to oxidative stress [
17]. The
agr quorum-sensing system response regulator, AgrA, was one of them.
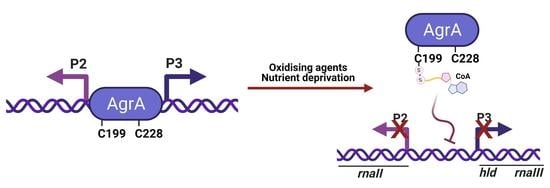
In this study, we report covalent modification of AgrA by CoA in vitro and in vivo, as well as the consequent regulation of its DNA-binding activity. External challenges in the form of oxidative stress and nutrient deprivation were found to induce AgrA CoAlation. The modified cysteines residues were mapped by liquid chromatography tandem mass spectrometry (LC–MS/MS) to Cys6 and Cys199, the latter of which is located in the LytTR DNA-binding domain. Furthermore, surface plasmon resonance (SPR) showed that in vitro CoAlation of recombinant AgrA inhibits its binding to both the P2 and P3 promoters. On the basis of these findings, we propose that CoAlation of AgrA at Cys199 under oxidative or metabolic stress modulates its DNA-binding activity and may create a binding motif for the formation of novel regulatory complexes implicated in the oxidative stress response. CoAlation of AgrA thus suggests an elaborate mechanism to regulate AgrA activity that synchronizes inputs from diverse stress response pathways to calibrate quorum sensing with other environmental or intracellular stimuli.
2. Materials and Methods
2.1. Reagents and Chemicals
All reagents and chemicals were obtained from Sigma-Aldrich unless stated otherwise.
2.2. Bacterial Growth Conditions, SaAgrA Overexpression and Treatmeants
S. aureus (MRSA: DSM11729) and Escherichia coli (E. coli) BL21 (DE3) cells transformed with pET28b(+)/ His-SaAgrA were cultured overnight in Luria–Bertani (LB) medium. The overnight cultures were diluted 1:100 in the same medium and incubated until the optical density at 600 nm reached 0.7 (OD600 = 0.7). For 6xHis-SaAgrA overexpression, pET28b(+)/His-SaAgrA transformed E. coli cells were induced with 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 20 min at 37 °C. To induce oxidative stress, cells were treated with or without oxidizing agents for 30 min at 37 °C.
For glucose-deprivation induced stress, E. coli BL21 (DE3) cells transformed with pET28b(+)/His-SaAgrA were induced with IPTG, harvested by centrifugation, resuspended in M9 minimal medium without glucose to remove the source of carbohydrate, and cultured at 37 °C for 30 min. For recovery experiments, starved cells were harvested and resuspended in M9 media supplemented with 20 mM glucose and incubated at 37 °C for 30 min or 60 min.
For nitrogen starvation experiments, E. coli BL21 (DE3) cells transformed with pET28b(+)/His-SaAgrA were grown in 10 mM NH4Cl (for overnight cultures) or 3 mM NH4Cl Gutnick minimal medium (for nitrogen starvation experiments), consisting of 33.8 mM KH2PO4, 77.5 mM K2HPO4, 5.74 mM K2SO4, and 0.41 mM MgSO4, supplemented with Ho-LE trace elements and 0.4% (wt/vol) glucose, using NH4Cl as the sole nitrogen source. NH4Cl concentration in the media was determined using the Aquaquant ammonium quantification kit (Merck Millipore), according to the manufacturer’s instructions. The time when the ammonium ran out ([ammonium] < 0.000625 mM) in the growth medium was used as a starting point for 30 min and 60 min incubation at 37 °C for the induction of the nitrogen starvation stress. For recovery experiments, starved cells were harvested by centrifugation and resuspended in 10 mM NH4Cl supplemented-Gutnick minimal medium and incubated for 30 min or 60 min at 37 °C.
2.3. Lysis of Cells, Protein Extraction and Affinity Purification
To extract proteins, harvested E. coli and S. aureus cells were resuspended in cell lysis buffer containing 20 mM Tris–HCl (pH 7.5), 50 mM NaCl, 50 mM NaF, 5 mM Na4P2O7, 0.5 mg/mL lysozyme, 100 mM N-Ethylmaleimide (NEM), and a cocktail of protease inhibitors (Roche). Sodium dodecyl sulfate (SDS) was added (1% final), and the homogenate was sonicated (30s on, 20 s off, 5 cycles) at 4 °C to reduce viscosity before centrifuging at 21,000× g for 10 min at 4 °C. The supernatant was collected and samples were boiled in 1X non-reducing SDS loading buffer (63 mM Tris HCl pH 8.0, 10% glycerol, 2% SDS, 0.0025% bromophenol blue) for 5 min before SDS-PAGE analysis and Western blotting (WB).
The affinity purification of 6xHis-SaAgrA from E. coli cells in oxidative stress and carbon or nitrogen deprivation experiments was carried out by incubating each sample with 10 µL bead volume of Ni2+ nitrilotriacetic acid (Ni-NTA) Sepharose beads for 30 min at 4 °C. Beads were washed three times with wash buffer (20 mM Tris HCl (pH 7.5), 50 mM NaCl, 50 mM NaF, 5 mM Na4P2O7) by centrifugation at 1000× g (4 °C, 2 min each). Beads were boiled in 1X non-reducing SDS loading dye for 5 min before analysis via WB.
2.4. Western Blot (WB) Analysis
Samples of bacterial extracts containing 30–40 µg of proteins, or total protein samples eluted from Ni-NTA beads were separated by SDS–polyacrylamide gel electrophoresis (PAGE) on 4–20% Mini-PROTEAN TGX Precast Gels (Bio-Rad Laboratories, Hercules, CA, USA). Separated proteins were transferred from the gel to a low-fluorescence polyvinylidene fluoride membrane (Bio-Rad Laboratories), which was then blocked with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE, USA). Mouse monoclonal anti-CoA antibody (0.17 µg/mL, generated as described previously [
20]) and rabbit polyclonal anti-AgrA (WB dilution 1:250, Eurogentech, Liège, Belgium) were diluted in Odyssey blocking buffer and incubated with the membrane for 2h at RT or overnight at 4 °C. Immunoreactive protein bands were visualized using Alexa Fluor 680 goat anti-mouse IgG H&L (WB dilution 1:10,000, Life Technologies, Carlsbad, CA, USA) and IRdye 800 CW goat anti-rabbit IgG H&L (WB dilution 1:10,000, LI-COR Biosciences) on the Odyssey infrared imaging system (Odyssey Scanner CLx and Image Studio Lite software, LI-COR Biosciences). For quantitative analysis, the band intensity values for CoAlated
SaAgrA were obtained from anti-CoA or anti-AgrA WB through Image Studio Lite (Ver 5.2). The anti-CoA band intensities were normalized against the corresponding anti-AgrA band intensities for each WB. The mean fold increase in CoAlation signal was calculated by comparison to respective controls. For statistical analysis, a ratio paired, one-tailed Student’s
t-test was used with GraphPad Prism (Version 9.1.1). Statistical significance was established for
p < 0.05, and the statistical variability was estimated with the standard error of the mean (SEM).
2.5. Sample Preparation for Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
Proteins from prepared cell lysates of diamide-treated
S. aureus cells were precipitated with 90% methanol. The protein pellet was resuspended in 50 mM ammonium bicarbonate (Ambic, pH 7.8) supplemented with 6.4 mM iodoacetamide (IAM), digested with endoproteinases Lys C and trypsin (sequencing grade, Promega, Madison, WI, USA) and heat inactivated at 99 °C, 10 min. CoAlated peptides were then immunoprecipitated with anti-CoA antibody cross-linked to Protein G Sepharose. Immunoprecipitated peptide mixtures were eluted with 0.1% trifluoroacetic acid (TFA), dried down completely in a SpeedVac and resolubilized in 20 µL of 50 mM Ambic and treated with 1.7 µg Nudix 7 phosphatase in the presence of 5 mM MgCl
2 at 37 °C for 20 min. Then the samples were acidified, desalted with a C18 Stage tip that contained 1.5 µL of Poros R3 resin and partially dried in a SpeedVac. Desalted peptides were further incubated for 45 min with 30 µL of Phos-Select IMAC resin (Sigma) in 100 µL of 30% MeCN, 0.25 M acetic acid (loading solution) for enrichment. Beads were washed four times with loading solution and CoAlated peptides were eluted twice with 500 mM imidazole (pH 7.6) and once with 30% MeCN/500 mM imidazole (pH 7.6). Before mass spectrometry analysis, CoAlated peptides were acidified, dried, desalted and partially dried using a SpeedVac. LC-MS/MS analysis and identification of CoAlated peptides from diamide-treated
S. aureus cells were carried out as previously described in [
17].
2.6. Expression and Purification of Recombinant 6xHis-SaAgrA Protein
The
S. aureus agrA coding sequence was cloned into the pET28b expression vector, and the recombinant protein was overexpressed in
E. coli Rosetta (DE3) pLysS cells [
21]. Briefly, cells were grown in LB at 37 °C to an optical density of 0.5 at 600 nm (OD
600). Subsequently, the expression of 6xHis-
SaAgrA was induced with 0.5 mM IPTG. The induced cells were grown for 16 h at 18 °C and harvested by centrifugation at 1000×
g. The harvested cells were suspended in a buffer containing 20 mM HEPES (pH 7.6), 300 mM KCl, 10% glycerol, and 2 mM phenylmethylsulfonyl fluoride (PMSF). After sonication (3 s on, 5 s off, 3 min, 2 cycles), the cell debris was separated by centrifugation at 8000×
g for 45 min at 4 °C. The cell-free lysate was then incubated with Ni-NTA Sepharose beads (Sigma-Aldrich, Inc., St. Louis, MO, USA) for 45 min at 4 °C. The bound protein was eluted in a buffer containing 20 mM HEPES (pH 7.6), 300 mM KCl, 10% glycerol, and 250 mM imidazole. The partially purified protein fractions were concentrated and loaded onto a Sephacryl S-200 (HiPrep 16/60) column (GE Healthcare, Chicago, IL, USA) equilibrated with 20 mM HEPES (pH 7.6), 250 mM KCl, and 10% glycerol for further purification by size exclusion chromatography. The purity of the sample was analyzed on a 12% SDS-PAGE gel, and the concentration was estimated using Bradford reagent (BioRad, Inc., Hercules, CA, USA) (
Figure S1).
2.7. In Vitro CoAlation of AgrA
A total of 20 μL of reduced AgrA (40 μM) was incubated with 5 μL of 600 μM coenzyme A (CoA, C3019- Sigma Aldrich) for 10 min to allow CoA binding. Subsequently, 5 μL of 1.8 mM H2O2 was added to the reaction mixture and incubated for half an hour at 25 °C. The reaction mixture was passed through a MicroBiospinTM 6 column to remove excess CoA and H2O2 before SPR analysis or treated with 10 mM NEM for 5 min at RT before WB analysis with anti-CoA antibody.
2.8. Surface Plasmon Resonance (SPR)
Interaction studies of AgrA and CoAlated AgrA with P2 and P3 promoter DNA were performed on a BIACORE 2000 instrument (Biacore, Uppsala, Sweden). Both P2 and P3 promoters (complementary sequences 5′ TAACAGTTAAGTATTTATTTCCTACAGTTAGGCA 3′ and 5′ TTCTTAACTAGTCGTTTTTTATTCTTAACTGTAA 3′, respectively) were biotinylated at the 5′ end (Sigma Aldrich, Co.) and annealed with the unlabeled complementary strand prior to immobilization on a Streptavidin (SA) sensor chip (GE Healthcare). For annealing, an equimolar concentration of labelled and unlabeled DNA strands was dissolved in 1X Saline Sodium Citrate (1X SSC) buffer and denatured at 100 °C for 10 min in a water bath and gradually annealed to room temperature. A total of 1 μM of the hybridized promoters were immobilized on the flow cells (P1 in channel 2, P2 in channel 3) using 10 mM sodium acetate (pH 4.0), and the chip was primed with 1X PBST (1X PBS with 0.05% Tween 20). Approximately 500 RUs (response difference units) of each promoter were immobilized. The first flow cell (channel 1) in the sensor chip was used as the reference. The interaction experiments were performed in a flow buffer containing 20 mM HEPES (pH 7.6), 250 mM KCl, and 10% glycerol. A total of 50 µL of the substrate (AgrA or CoAlated AgrA) at various concentrations were passed over the flow cells (flow rate: 30 µL/min) and allowed to dissociate for 200 s. The sensor surface was regenerated with multiple injections of 0.05–0.1% SDS whenever required. The normalized response curves (reference subtracted) obtained were evaluated using BIA evaluation software. The data obtained were fit to a Langmuir 1:1 interaction model to obtain the rates of association (K
a); dissociation (K
d); and the equilibrium dissociation constant, K
D (K
d/K
a) (
Table 1). The consistency between multiple datasets (performed with different protein preparations) was evaluated by comparing the values of theoretically fitted K
d and calculated K
d using BIA evaluation software.
4. Discussion
Bacterial cells use various strategies to maintain redox homeostasis, including transcriptional regulation, which allows for the expression of genes involved in antioxidant defense [
26]. Redox-sensing regulators in bacteria respond to diverse environmental cues such as the availability of nutrients and oxygen, as well as exposure to reactive oxygen and nitrogen species, among others. These redox signals are transduced by transcriptional regulators through specific mechanisms, involving upregulated expression of low molecular weight thiols, antioxidant enzymes, and detoxifying proteins [
27,
28,
29].
S. aureus is an aggressive opportunistic pathogen due to its prominent virulence and antibiotic resistance, which is achieved through adaptive and timely coordination of gene expression for virulence, growth, and survival [
2]. These include two-component regulatory systems, transcription factors, and regulatory RNAs.
S. aureus expresses over 250 sRNA genes, many of which are responsible for the adaptation to environmental changes including oxidative and metabolic stress conditions. The
agr two-component system is perhaps the most-studied and has well elucidated roles in quorum-sensing; however, much less is known about its role in oxidation sensing [
1].
The recent development of an LC–MS/MS methodology and a specific anti-CoA antibody allowed for a proteome-wide CoAlome analysis in mammalian and bacterial cells, which identified over 2100 CoAlated proteins in response to oxidative or metabolic stress. Functional classification of CoAlated proteins revealed that in contrast to mammalian cells, numerous transcription factors and regulators are found to be CoA-modified in bacteria [
16,
17]. The susceptibility of bacterial transcription factors to oxPTMs suggests their importance in the antioxidant response. The most studied oxPTM is glutathionylation, and it was previously shown to be involved in the modulation of bacterial virulence and the activity of transcriptional regulators [
9,
10]. However, GSH is not available as an antioxidant in
S. aureus, which only produces two low molecular weight thiols, BSH and CoA. BSH is considered a key protective thiol in
S. aureus antioxidant defense by forming protein–BSH mixed disulfides through bacillithiolation [
30,
31]. While there are no reports to our knowledge describing bacillithiolation of AgrA during oxidative stress, it has been found that AgrA possesses a redox-sensitive cysteine residue that is a target of oxidative stress response, and therefore susceptible to oxPTMs. Cys199, located in the DNA-binding domain of AgrA, was identified as the oxidation-sensing residue, and its oxidation was shown to inhibit the DNA-binding activity of AgrA [
5]. Furthermore, another study revealed that Cys199 of
S. aureus AgrA (among other cysteine residues) is a substrate for S-nitrosylation, and this modification was shown to inhibit
agr transcription [
32].
In the present study, we reveal the induction of AgrA CoAlation in response to a panel of oxidizing agents (diamide, H
2O
2, and TBH) and metabolic stress induced by glucose or nitrogen deprivation. The sites of CoAlation were mapped to Cys199 and Cys6 using LC–MS/MS. Since Cys199 is the redox-sensitive residue located in the DNA-binding region, we hypothesized that CoAlation could interfere with DNA binding. Indeed, SPR analysis showed that CoA-modified AgrA had significantly lower affinity towards the P2 and P3 promoters than non-CoAlated AgrA. Active AgrA triggers transcription from its own operon (agrBDCA, i.e., the P2 promoter), as well as the divergently transcribed regulatory RNAIII (i.e., the P3 promoter). Dissociation of CoAlated AgrA from its promoters would therefore result in decreased transcription of AgrB, AgrD, AgrC, and AgrA, as well as the downregulation of RNAIII expression (
Figure 6).
It was previously shown in a microarray study that active AgrA downregulates transcription of the
S. aureus GSH peroxidase gene encoding for
BsaA, a key enzyme in bacterial resistance to oxidative stress [
33]. Another study confirmed that AgrA represses
BsaA expression, suggesting that it occurs via a direct DNA-binding mechanism [
5]. Interestingly, oxidative stress was shown to relieve the AgrA-mediated downregulation of
BsaA expression. Similarly, oxidative- or metabolic stress-induced CoAlation of AgrA might result in de-repression of
BsaA transcription and allow for efficient bacterial antioxidant defense. Dissociation of CoAlated AgrA from the P2 promoter might constitute an adaptive response in bacteria to initiate antioxidant defense, revealing the protective role of CoAlation in
S. aureus.
Protein CoAlation was described as a reversible PTM in bacteria and mammalian cells where numerous proteins were shown to be efficiently deCoAlated after the removal of oxidizing agents or metabolic stress [
16,
17]. Data presented in this study reveal that AgrA CoAlation is reversible after the recovery of cells from glucose starvation-induced metabolic stress, which was also the case for numerous proteins in the total cell lysates (
Figure 3). This is consistent with previous findings which showed that protein CoAlation in bacteria (
E. coli,
B. megaterium, and
S. aureus) exposed to oxidative or metabolic stress is efficiently reversed after the recovery of cells in oxidant-free media. Previous studies described the effects of CoAlation on metabolic enzymes in bacteria and mammalian cells. Numerous enzymes including creatine kinase, peroxiredoxin 5, GAPDH, and Aurora A kinase were found to be CoAlated in response to oxidative or metabolic stress, and CoAlation was shown to affect their activity and protect them from irreversible overoxidation [
19,
34,
35]. It was also proposed that CoAlation might serve as a scaffold for the formation of regulatory interactions and complexes. Since CoA is a relatively large molecule comprising of a pantetheine tail and an ADP moiety, protein CoAlation might form novel binding sites, specifically for proteins containing the Rossmann fold that could recognize the ADP moiety of CoA. In the case of AgrA, covalent modification by CoA under oxidative or metabolic stress may allow for the formation of regulatory interactions implicated in the transduction of redox signaling and antioxidant gene expression (
Figure 6).