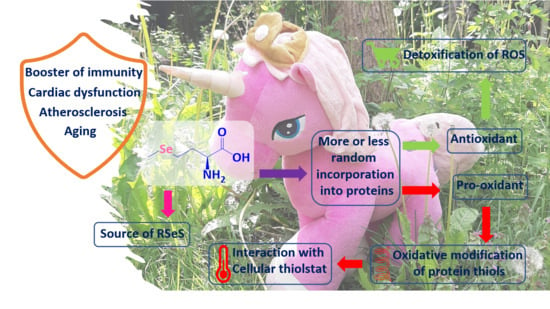
Selenomethionine: A Pink Trojan Redox Horse with Implications in Aging and Various Age-Related Diseases
Abstract
:1. Introduction
2. Biosynthesis of SeMet in Plants
3. SeMet in Yeast
4. Redox Activity and Catalysis of SeMet
5. Implications of SeMet in Aging and Diseases Related to OS
5.1. Selenomethionine Supplements Minimize Cardiac Dysfunction
5.2. SeMet Regulates Inflammatory Reactions in Atherosclerosis
5.3. SeMet and Viral Infections
5.4. SeMet and Cancer
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernández-Lázaro, D.; Fernandez-Lazaro, C.I.; Mielgo-Ayuso, J.; Navascués, L.J.; Martínez, A.C.; Seco-Calvo, J. The Role of Selenium Mineral Trace Element in Exercise: Antioxidant Defense System, Muscle Performance, Hormone Response, and Athletic Performance. A Systematic Review. Nutrients 2020, 12, 1790. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Sarma, B.K.; Phadnis, P.P.; Mugesh, G. Selenium-containing enzymes in mammals: Chemical perspectives. J. Chem. Sci. 2005, 117, 287–303. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, R.; Hasegawa, M.; Kawaguchi, C.; Ishikawa, N.; Tomiwa, K.; Shima, M.; Nogami, K. Thyroid function in patients with selenium deficiency exhibits high free T4 to T3 ratio. Clin. Pediatr. Endocrinol. 2021, 30, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Yabu, T.; Yamashita, M. Discovery of the strong antioxidant selenoneine in tuna and selenium redox metabolism. World J. Biol. Chem. 2010, 1, 144–150. [Google Scholar] [CrossRef]
- Leblanc, K.L.; Smith, M.S.; Wallschläger, D. Production and Release of Selenocyanate by Different Green Freshwater Algae in Environmental and Laboratory Samples. Environ. Sci. Technol. 2012, 46, 5867–5875. [Google Scholar] [CrossRef] [PubMed]
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Liang, X.; Fomenko, D.E.; Raza, A.S.; Chong, C.-K.; Carlson, B.A.; Hatfield, D.L.; Gladyshev, V.N. Analysis of Methionine/Selenomethionine Oxidation and Methionine Sulfoxide Reductase Function Using Methionine-Rich Proteins and Antibodies against Their Oxidized Forms†. Biochemistry 2008, 47, 6685–6694. [Google Scholar] [CrossRef] [Green Version]
- Burk, R.F. Selenium: Recent clinical advances. Curr. Opin. Gastroenterol. 2001, 17, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, S.; Dharmaraj, S. Selenium and selenoproteins: It’s role in regulation of inflammation. Inflammopharmacology 2020, 28, 667–695. [Google Scholar] [CrossRef]
- Solovyev, N.; Drobyshev, E.; Blume, B.; Michalke, B. Selenium at the Neural Barriers: A Review. Front. Neurosci. 2021, 15. [Google Scholar] [CrossRef]
- Kang, D.; Lee, J.; Wu, C.; Guo, X.; Lee, B.J.; Chun, J.-S.; Kim, J.-H. The role of selenium metabolism and selenoproteins in cartilage homeostasis and arthropathies. Exp. Mol. Med. 2020, 52, 1198–1208. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, X.; Ma, J.; Mu, Y.; Wang, Y.; Yang, S.; Wu, Y.; Wu, F.; Zhou, Y. Selenium (Se) plays a key role in the biological effects of some viruses: Implications for COVID-19. Environ. Res. 2021, 196, 110984. [Google Scholar] [CrossRef]
- Hou, L.; Lin, Z.; Xu, A.; Le, G.; Ge, L.; Liu, S.; Muhmood, A.; Gan, F.; Huang, K. Combined protective effects of icariin and selenomethionine on novel chronic tubulointerstitial nephropathy models in vivo and in vitro. Br. J. Nutr. 2021, 1–33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Falandysz, J. Selenium in Edible Mushrooms. J. Environ. Sci. Health Part C 2008, 26, 256–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimaszewska, M.; Górska, S.; Dawidowski, M.; Podsadni, P.; Turło, J. Biosynthesis of Se-methyl-seleno-l-cysteine in Basidiomycetes fungus Lentinula edodes (Berk.) Pegler. SpringerPlus 2016, 5, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; McGrath, S.P.; Zhao, F. Selenium uptake, translocation and speciation in wheat supplied with selenate or selenite. New Phytol. 2008, 178, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Q.; Mitani, N.; Yamaji, N.; Shen, R.F.; Ma, J.F. Involvement of Silicon Influx Transporter OsNIP2;1 in Selenite Uptake in Rice. Plant Physiol. 2010, 153, 1871–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Su, Y.; Li, L.; Huang, X.; Panhwar, F.H.; Zheng, T.; Tang, Z.; Ei, H.H.; Farooq, M.U.; Zeng, R.; et al. Quick selenium accumulation in the selenium-rich rice and its physiological responses in changing selenium environments. BMC Plant Biol. 2019, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- White, P.J. Selenium accumulation by plants. Ann. Bot. 2016, 117, 217–235. [Google Scholar] [CrossRef] [Green Version]
- Winkel, L.H.; Vriens, B.; Jones, G.D.; Schneider, L.S.; Pilon-Smits, E.; Bañuelos, G.S. Selenium Cycling Across Soil-Plant-Atmosphere Interfaces: A Critical Review. Nutrients 2015, 7, 4199–4239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, B.; Anderson, J.W. Synthesis of selenocysteine by cysteine synthases from selenium accumulator and non-accumulator plants. Phytochemistry 1978, 17, 2069–2074. [Google Scholar] [CrossRef]
- Van Hoewyk, D.; Çakir, O. Manipulating Selenium Metabolism in Plants: A Simple Twist of Metabolic Fate Can Alter Selenium Tolerance and Accumulation. In Plant Ecophysiology; Pilon-Smits, E.A.H., Winkel, L.H.E., Lin, Z.-Q., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 11, pp. 165–176. [Google Scholar]
- Zhou, Z.S.; Smith, A.E.; Matthews, R.G. l-selenohomocysteine: One-step synthesis from l-selenomethionine and kinetic analysis as substrate for methionine synthases. Bioorg. Med. Chem. Lett. 2000, 10, 2471–2475. [Google Scholar] [CrossRef]
- Gajda, J.; Potrzebowski, M.J.; Bujacz, A.; Bujacz, G. Application of the 77Se Solid State NMR for Investigation of Bioorganic Compounds—The Case of Selenomethionine. Phosphorus Sulfur Silicon Relat. Elements 2008, 183, 1061–1066. [Google Scholar] [CrossRef]
- Al Ghanem, A.; Nasim, M.J.; Alnahas, F.; Ney, Y.; Weiss, A.-V.; Koch, M.; Schneider, M.; Jacob, C. Incredible edible selenium nanoparticles produced by food-grade microorganisms. Curr. Nutraceuticals 2020, 1, 1–10. [Google Scholar] [CrossRef]
- Herrero, E.; Wellinger, R.E. Yeast as a model system to study metabolic impact of selenium compounds. Microb. Cell 2015, 2, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Hahn, G.A.; Brown, J.W. Properties of a methionyl-tRNA systhetase from Sarcina lutea. Biochim. Biophys. Acta (BBA) Enzym. 1967, 146, 264–271. [Google Scholar] [CrossRef]
- Kitajima, T.; Chiba, Y. Selenomethionine metabolism and its toxicity in yeast. Biomol. Concepts 2013, 4, 611–616. [Google Scholar] [CrossRef]
- Chiao, J.S.; Peterson, W.H. Yeasts, Methionine and Cystine Contents. J. Agric. Food Chem. 1953, 1, 1005–1008. [Google Scholar] [CrossRef]
- Ouerdane, L.; Mester, Z. Production and Characterization of Fully Selenomethionine-Labeled Saccharomyces cerevisiae. J. Agric. Food Chem. 2008, 56, 11792–11799. [Google Scholar] [CrossRef]
- Encinar, J.R.; Ouerdane, L.; Buchmann, W.; Tortajada, J.; Lobinski, R.; Szpunar, J. Identification of Water-Soluble Selenium-Containing Proteins in Selenized Yeast by Size-Exclusion-Reversed-Phase HPLC/ICPMS Followed by MALDI-TOF and Electrospray Q-TOF Mass Spectrometry. Anal. Chem. 2003, 75, 3765–3774. [Google Scholar] [CrossRef]
- Rao, Y.; Mc Cooeye, M.; Windust, A.; Bramanti, E.; D’Ulivo, A.; Mester, Z. Mapping of Selenium Metabolic Pathway in Yeast by Liquid Chromatography−Orbitrap Mass Spectrometry. Anal. Chem. 2010, 82, 8121–8130. [Google Scholar] [CrossRef] [Green Version]
- Craddock, V.M. Reactivity of selenomethionine in nucleic acid methylase reactions in the rat. Chem. Interact. 1972, 5, 207–211. [Google Scholar] [CrossRef]
- Kitajima, T.; Jigami, Y.; Chiba, Y. Cytotoxic Mechanism of Selenomethionine in Yeast*. J. Biol. Chem. 2012, 287, 10032–10038. [Google Scholar] [CrossRef] [Green Version]
- Miranda, S.; Wang, Y.; Purdie, N.; Osborne, V.; Coomber, B.; Cant, J. Selenomethionine stimulates expression of glutathione peroxidase 1 and 3 and growth of bovine mammary epithelial cells in primary culture. J. Dairy Sci. 2009, 92, 2670–2683. [Google Scholar] [CrossRef] [Green Version]
- Briviba, K.; Roussyn, I.; Sharov, V.S.; Sies, H. Attenuation of oxidation and nitration reactions of peroxynitrite by selenomethionine, selenocystine and ebselen. Biochem. J. 1996, 319, 13–15. [Google Scholar] [CrossRef] [Green Version]
- Roussyn, I.; Briviba, K.; Masumoto, H.; Sies, H. Selenium-Containing Compounds Protect DNA fromSingle-Strand Breaks Caused by Peroxynitrite. Arch. Biochem. Biophys. 1996, 330, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Padmaja, S.; Squadrito, G.; Lemercier, J.-N.; Cueto, R.; Pryor, W.A. Rapid oxidation of dl-selenomethionine by peroxynitrite. Free Radic. Biol. Med. 1996, 21, 317–322. [Google Scholar] [CrossRef]
- Alvarez, B.; Ferrer-Sueta, G.; Freeman, B.A.; Radi, R. Kinetics of Peroxynitrite Reaction with Amino Acids and Human Serum Albumin. J. Biol. Chem. 1999, 274, 842–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, R.J.; Glocke, S.C.; Sicuri, A.R.; Ripp, A.S.L.; Elfarra, A.A. Oxidative Metabolism of Seleno-l-methionine tol-Methionine Selenoxide by Flavin-Containing Monooxygenases. Chem. Res. Toxicol. 2006, 19, 1643–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahmanto, A.S.; Davies, M.J. Catalytic activity of selenomethionine in removing amino acid, peptide, and protein hydroperoxides. Free Radic. Biol. Med. 2011, 51, 2288–2299. [Google Scholar] [CrossRef]
- Rahmanto, A.S.; Davies, M.J. Selenium-containing amino acids as direct and indirect antioxidants. IUBMB Life 2012, 64, 863–871. [Google Scholar] [CrossRef]
- Skaff, O.; Pattison, D.I.; Morgan, P.E.; Bachana, R.; Jain, V.K.; Priyadarsini, K.I.; Davies, M.J. Selenium-containing amino acids are targets for myeloperoxidase-derived hypothiocyanous acid: Determination of absolute rate constants and implications for biological damage. Biochem. J. 2011, 441, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Davis, S.J.; Ikemizu, S.; Collins, A.V.; Fennelly, J.A.; Harlos, K.; Jones, E.Y.; Stuart, D. Crystallization and functional analysis of a soluble deglycosylated form of the human costimulatory molecule B7-1. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001, 57, 605–608. [Google Scholar] [CrossRef] [Green Version]
- Barton, W.A.; Tzvetkova-Robev, R.; Erdjument-Bromage, H.; Tempst, P.; Nikolov, D.B. Highly efficient selenomethionine labeling of recombinant proteins produced in mammalian cells. Protein Sci. 2006, 15, 2008–2013. [Google Scholar] [CrossRef] [Green Version]
- Hatfield, D.L.; Gladyshev, V.N. How Selenium Has Altered Our Understanding of the Genetic Code. Mol. Cell. Biol. 2002, 22, 3565–3576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, L.; Pattison, D.I.; Fu, S.; Schiesser, C.H.; Davies, M.J.; Hawkins, C.L. Catalytic oxidant scavenging by selenium-containing compounds: Reduction of selenoxides and N-chloramines by thiols and redox enzymes. Redox Biol. 2017, 12, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.C.; Le, D.T.; Gladyshev, V.N. Mammals Reduce Methionine-S-sulfoxide with MsrA and Are Unable to Reduce Methionine-R-sulfoxide, and This Function Can Be Restored with a Yeast Reductase. J. Biol. Chem. 2008, 283, 28361–28369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrauzer, G.N. The nutritional significance, metabolism and toxicology of selenomethionine. Adv. Food Nutr. Res. 2003, 47, 73–112. [Google Scholar] [CrossRef]
- Peoples, J.N.; Saraf, A.; Ghazal, N.; Pham, T.T.; Kwong, J.Q. Mitochondrial dysfunction and oxidative stress in heart disease. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Park, M.; Kang, C.; Dilmen, S.; Kang, T.H.; Kang, D.G.; Ke, Q.; Lee, S.U.; Lee, D.; Kang, P.M. Hydrogen Peroxide-Responsive Nanoparticle Reduces Myocardial Ischemia/Reperfusion Injury. J. Am. Heart Assoc. 2016, 5, e003697. [Google Scholar] [CrossRef] [PubMed]
- Reyes, L.; Bishop, D.P.; Hawkins, C.L.; Rayner, B.S. Assessing the Efficacy of Dietary Selenomethionine Supplementation in the Setting of Cardiac Ischemia/Reperfusion Injury. Antioxidants 2019, 8, 546. [Google Scholar] [CrossRef] [Green Version]
- Karaye, K.M.; Sa’Idu, H.; Balarabe, S.A.; Ishaq, N.A.; Sanni, B.; Abubakar, H.; Mohammed, B.L.; Abdulsalam, T.; Tukur, J.; Mohammed, I.Y. Selenium supplementation in patients with peripartum cardiomyopathy: A proof-of-concept trial. BMC Cardiovasc. Disord. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Tyrrell, D.J.; Goldstein, D.R. Ageing and atherosclerosis: Vascular intrinsic and extrinsic factors and potential role of IL-6. Nat. Rev. Cardiol. 2021, 18, 58–68. [Google Scholar] [CrossRef]
- Hu, Y.; Lee, S.O.; Lou, W.; Gao, A.C. Down-Regulation of IL-6 by Selenium in the Androgen-Independent Prostate Cancer. Cancer Res. 2007, 67, 315. [Google Scholar]
- Prystupa, A.; Kiciński, P.; Luchowska-Kocot, D.; Błażewicz, A.; Niedziałek, J.; Mizerski, G.; Jojczuk, M.; Ochal, A.; Sak, J.J.; Załuska, W. Association between Serum Selenium Concentrations and Levels of Proinflammatory and Profibrotic Cytokines—Interleukin-6 and Growth Differentiation Factor-15, in Patients with Alcoholic Liver Cirrhosis. Int. J. Environ. Res. Public Health 2017, 14, 437. [Google Scholar] [CrossRef]
- Liu, H.; Xu, H.; Huang, K. Selenium in the prevention of atherosclerosis and its underlying mechanisms. Metallomics 2017, 9, 21–37. [Google Scholar] [CrossRef]
- Zhang, Y.; Cartland, S.P.; Henriquez, R.; Patel, S.; Gammelgaard, B.; Flouda, K.; Hawkins, C.; Rayner, B.S. Selenomethionine supplementation reduces lesion burden, improves vessel function and modulates the inflammatory response within the setting of atherosclerosis. Redox Biol. 2020, 29, 101409. [Google Scholar] [CrossRef] [PubMed]
- Shojadoost, B.; Kulkarni, R.R.; Yitbarek, A.; Laursen, A.; Taha-Abdelaziz, K.; Alkie, T.N.; Barjesteh, N.; Quinteiro-Filho, W.M.; Smith, T.K.; Sharif, S. Dietary selenium supplementation enhances antiviral immunity in chickens challenged with low pathogenic avian influenza virus subtype H9N2. Vet. Immunol. Immunopathol. 2019, 207, 62–68. [Google Scholar] [CrossRef]
- Zhang, J.; Taylor, E.W.; Bennett, K.; Saad, R.; Rayman, M.P. Association between regional selenium status and reported outcome of COVID-19 cases in China. Am. J. Clin. Nutr. 2020, 111, 1297–1299. [Google Scholar] [CrossRef] [PubMed]
- Khatiwada, S.; Subedi, A. A Mechanistic Link Between Selenium and Coronavirus Disease 2019 (COVID-19). Curr. Nutr. Rep. 2021, 10, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Cermelli, C.; Vinceti, M.; Scaltriti, E.; Bazzani, E.; Beretti, F.; Vivoli, G.; Portolani, M. Selenite inhibition of Coxsackie virus B5 replication: Implications on the etiology of Keshan disease. J. Trace Elem. Med. Biol. 2002, 16, 41–46. [Google Scholar] [CrossRef]
- Allan, G.M.; McNeilly, E.; Kennedy, S.; Meehan, B.; Moffett, D.; Malone, F.; Ellis, J.; Krakowka, S. PCV-2-associated PDNS in Northern Ireland in 1990. Porcine dermatitis and nephropathy syndrome. Vet. Rec. 2000, 146, 711–712. [Google Scholar]
- Ficek, R.; Pšikal, I.; Fictum, P.; Bendová, J.; Kosinová, E.; Smítalová, R.; Škorič, M. Exploratory Epidemiological Study on Porcine Circovirus Type 2 Infection and Postweaning Multisystemic Wasting Syndrome in the Czech Republic. Acta Vet. Brno 2010, 79, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Stevenson, G.W.; Kiupel, M.; Harrach, B.; Anothayanontha, L.; Kanitz, C.L.; Mittal, S.K. Sequence Analysis of Old and New Strains of Porcine Circovirus Associated with Congenital Tremors in Pigs and Their Comparison with Strains Involved with Postweaning Multisystemic Wasting Syndrome Résumé. Can. J. Vet. Res. 2002, 66, 217–224. [Google Scholar] [PubMed]
- Laconi, E.; Marongiu, F.; DeGregori, J. Cancer as a disease of old age: Changing mutational and microenvironmental landscapes. Br. J. Cancer 2020, 122, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.M.; Jemal, A. Cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Fane, M.; Weeraratna, A.T. How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 2020, 20, 89–106. [Google Scholar] [CrossRef]
- Redman, C.; Scott, J.A.; Baines, A.T.; Basye, J.L.; Clark, L.C.; Calley, C.; Roe, D.; Payne, C.M.; Nelson, M.A. Inhibitory effect of selenomethionine on the growth of three selected human tumor cell lines. Cancer Lett. 1998, 125, 103–110. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, F.; Ren, Y.; Xing, L.; Wu, Y.; Li, Z.; Pan, H.; Xu, C. The Anticancer Effects of Sodium Selenite and Selenomethionine on Human Colorectal Carcinoma Cell Lines in Nude Mice. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2009, 18, 1–8. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Prabhu, K.S.; Das, A.; Mastro, A.M. Dietary selenium supplementation modifies breast tumor growth and metastasis. Int. J. Cancer 2013, 133, 2054–2064. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Stern, P.H.; Coalson, D.W.; Wallace, C.D.; Erbe, R.W. Altered Methionine Metabolism in Cancer Cells. Adv. Struct. Saf. Stud. 2019, 1866, 13–26. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasim, M.J.; Zuraik, M.M.; Abdin, A.Y.; Ney, Y.; Jacob, C. Selenomethionine: A Pink Trojan Redox Horse with Implications in Aging and Various Age-Related Diseases. Antioxidants 2021, 10, 882. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10060882
Nasim MJ, Zuraik MM, Abdin AY, Ney Y, Jacob C. Selenomethionine: A Pink Trojan Redox Horse with Implications in Aging and Various Age-Related Diseases. Antioxidants. 2021; 10(6):882. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10060882
Chicago/Turabian StyleNasim, Muhammad Jawad, Mhd Mouayad Zuraik, Ahmad Yaman Abdin, Yannick Ney, and Claus Jacob. 2021. "Selenomethionine: A Pink Trojan Redox Horse with Implications in Aging and Various Age-Related Diseases" Antioxidants 10, no. 6: 882. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10060882