Culture Conditions Affect Antioxidant Production, Metabolism and Related Biomarkers of the Microalgae Phaeodactylum tricornutum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation Conditions
2.2. Biomass Isolation
2.3. Lipid Content
Total Lipids and Fatty Acids
2.4. Antioxidant Assays
2.5. Real-Time PCR Analysis
2.6. Statistical Analysis
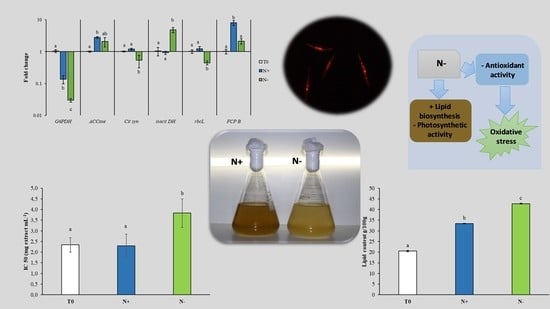
3. Results and Discussion
3.1. Cell Growth
3.2. Lipid Content
3.2.1. Total Lipid Content
3.2.2. Fatty Acid Content
3.3. Cellular Antioxidant Activity
3.3.1. Polyphenol Content
3.3.2. Carotenoids
3.3.3. Total Antioxidant Assays
3.4. Gene Expression
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, W.; Daboussi, F. Genetic and metabolic engineering in diatoms. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Ghosh, S.; Dubinsky, Z.; Verdelho, V.; Iluz, D. Unconventional high-value products from microalgae: A review. Bioresour. Technol. 2021, 329, 124895. [Google Scholar] [CrossRef] [PubMed]
- Andrade, D.S.; Amaral, H.F.; Gavilanes, F.Z.; Morioka, L.R.I.; Nassar, J.M.; de Melo, J.M.; Silva, H.R.; Telles, T.S. Microalgae: Cultivation, Biotechnological, Environmental, and Agricultural Applications. In Advances in the Domain of Environmental Biotechnology; Springer: Singapore, 2021; pp. 635–701. [Google Scholar]
- Maltsev, Y.; Maltseva, K. Fatty acids of microalgae: Diversity and applications. Rev. Environ. Sci. Bio/Technol. 2021, 20, 515–547. [Google Scholar] [CrossRef]
- Rizwan, M.; Mujtaba, G.; Ahmed, S.; Lee, K.; Rashid, N. Exploring the potential of microalgae for new biotechnology applications and beyond: A review. Renew. Sustain. Energy Rev. 2018, 92, 394–404. [Google Scholar] [CrossRef]
- Gallego-Cartagena, E.; Castillo-Ramírez, M.; Martínez-Burgos, W. Effect of stressful conditions on the carotenogenic activity of a Colombian strain of Dunaliella salina. Saudi J. Biol. Sci. 2019, 26, 1325–1330. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, N.T. The technology of microalgal culturing. Biotechnol. Lett. 2008, 30, 1525–1536. [Google Scholar] [CrossRef]
- Vigani, M.; Parisi, C.; Rodríguez-Cerezo, E.; Barbosa, M.J.; Sijtsma, L.; Ploeg, M.; Enzing, C. Food and feed products from micro-algae: Market opportunities and challenges for the EU. Trends Food Sci. Technol. 2015, 42, 81–92. [Google Scholar] [CrossRef]
- Xue, J.; Chen, T.T.; Zheng, J.W.; Balamurugan, S.; Liu, Y.H.; Yang, W.D.; Liu, J.S.; Li, H.Y. Glucose-6-Phosphate Dehydrogenase from the Oleaginous Microalga Nannochloropsis Uncovers Its Potential Role in Promoting Lipogenesis. Biotechnol. J. 2020, 15, 1900135. [Google Scholar] [CrossRef]
- Lafarga, T. Effect of microalgal biomass incorporation into foods: Nutritional and sensorial attributes of the end products. Algal Res. 2019, 41, 101566. [Google Scholar] [CrossRef]
- Ramesh Kumar, B.; Deviram, G.; Mathimani, T.; Duc, P.A.; Pugazhendhi, A. Microalgae as rich source of polyunsaturated fatty acids. Biocatal. Agric. Biotechnol. 2019, 17, 583–588. [Google Scholar] [CrossRef]
- Yarkent, Ç.; Gürlek, C.; Oncel, S.S. Potential of microalgal compounds in trending natural cosmetics: A review. Sustain. Chem. Pharm. 2020, 17, 100304. [Google Scholar] [CrossRef]
- Mishra, A.; Medhi, K.; Malaviya, P.; Thakur, I.S. Omics approaches for microalgal applications: Prospects and challenges. Bioresour. Technol. 2019, 291, 121890. [Google Scholar] [CrossRef] [PubMed]
- Yodsuwan, N.; Sawayama, S.; Sirisansaneeyakul, S. Effect of nitrogen concentration on growth, lipid production and fatty acid profiles of the marine diatom Phaeodactylum tricornutum. Agric. Nat. Resour. 2017, 51, 190–197. [Google Scholar] [CrossRef]
- Yang, Z.K.; Niu, Y.F.; Ma, Y.H.; Xue, J.; Zhang, M.H.; Yang, W.D.; Liu, J.S.; Lu, S.H.; Guan, Y.; Li, H.Y. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 2013, 6, 67. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yin, W.; Ma, D.; Liu, X.; Xu, K.; Liu, J. Phytohormone supplementation significantly increases fatty acid content of Phaeodactylum tricornutum in two-phase culture. J. Appl. Phycol. 2020, 33, 13–23. [Google Scholar] [CrossRef]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.; Chen, A.; Zhang, W.; Li, A.; Zhang, C. Co-production of lipids, eicosapentaenoic acid, fucoxanthin, and chrysolaminarin by Phaeodactylum tricornutum cultured in a flat-plate photobioreactor under varying nitrogen conditions. J. Ocean Univ. China 2017, 16, 916–924. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, M.K.; Li, C.Y.; Wang, Y.; Zhang, J.; Wang, D.B.; Zhang, X.E.; Ge, F. Phosphoproteomic analysis provides novel insights into stress responses in Phaeodactylum tricornutum, a model diatom. J. Proteome Res. 2014, 13, 2511–2523. [Google Scholar] [CrossRef] [PubMed]
- Krishna, A.; Wayne, K.; Rambabu, K.; Tao, Y.; Chu, D.; Show, P. Food Science and Human Wellness Microalgae: A potential alternative to health supplementation for humans. Food Sci. Hum. Wellness 2019, 8, 16–24. [Google Scholar]
- Yang, Z.K.; Ma, Y.H.; Zheng, J.W.; Yang, W.D.; Liu, J.S.; Li, H.Y. Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Appl. Phycol. 2014, 26, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.; Pienkos, P.T.; Pruthi, V.; Poluri, K.M.; Guarnieri, M.T. Leveraging algal omics to reveal potential targets for augmenting TAG accumulation. Biotechnol. Adv. 2018, 36, 1274–1292. [Google Scholar] [CrossRef]
- Fanesi, A.; Raven, J.A.; Giordano, M. Growth rate affects the responses of the green alga Tetraselmis suecica to external perturbations. Plant Cell Environ. 2014, 37, 512–519. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Sloane Stanley, G. A Simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1956, 55, 999–1033. [Google Scholar]
- Lepage, G.; Roy, C.C. Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J. Lipid Res. 1984, 25, 1391–1396. [Google Scholar] [CrossRef]
- Messina, C.M.; Renda, G.; La Barbera, L.; Santulli, A. By-products of farmed European sea bass (Dicentrarchus labrax L.) as a potential source of n-3 PUFA. Biologia 2013, 68, 288–293. [Google Scholar] [CrossRef] [Green Version]
- Bustos, R.; Romo, L.; Yáñez, K.; Díaz, G.; Romo, C. Oxidative stability of carotenoid pigments and polyunsaturated fatty acids in microparticulate diets containing krill oil for nutrition of marine fish larvae. J. Food Eng. 2003, 56, 289–293. [Google Scholar] [CrossRef]
- Safafar, H.; Van Wagenen, J.; Møller, P.; Jacobsen, C. Carotenoids, phenolic compounds and tocopherols contribute to the antioxidative properties of some microalgae species grown on industrial wastewater. Mar. Drugs 2015, 13, 7339–7356. [Google Scholar] [CrossRef] [Green Version]
- Folin, O.; Ciocalteu, V. Tyrosine and Tryptophane in Proteins. J. Biol. Chem. 1927, 73, 627–650. [Google Scholar] [CrossRef]
- Maadane, A.; Merghoub, N.; Ainane, T.; El Arroussi, H.; Benhima, R.; Amzazi, S.; Bakri, Y.; Wahby, I. Antioxidant activity of some Moroccan marine microalgae: Pufa profiles, carotenoids and phenolic content. J. Biotechnol. 2015, 215, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, 1, 431–438. [Google Scholar] [CrossRef]
- Bernatoniene, J.; Masteikova, R.; Davalgiene, J.; Peciura, R.; Gauryliene, R.; Bernatoniene, R.; Majiene, D.; Lazauskas, R.; Civinskiene, G.; Velziene, S.; et al. Topical application of Calendula officinalis (L.): Formulation and evaluation of hydrophilic cream with antioxidant activity. J. Med. Plants Res. 2011, 5, 868–877. [Google Scholar]
- Falleh, H.; Ksouri, R.; Medini, F.; Guyot, S.; Abdelly, C.; Magné, C. Antioxidant activity and phenolic composition of the medicinal and edible halophyte Mesembryanthemum edule L. Ind. Crops Prod. 2011, 34, 1066–1071. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, S.; Huang, A.; Zhang, B.; Huan, L.; Zhao, P.; Lin, A.; Wang, G. Enzyme activity highlights the importance of the oxidative pentose phosphate pathway in lipid accumulation and growth of Phaeodactylum tricornutum under CO2 concentration. Biotechnol. Biofuels 2015, 8, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef]
- Guerra, L.T.; Levitan, O.; Frada, M.J.; Sun, J.S.; Falkowski, P.G.; Dismukes, G.C. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy 2013, 59, 306–315. [Google Scholar] [CrossRef]
- Sachse, M.; Sturm, S.; Gruber, A.; Kroth, P. Identification and evaluation of endogenous reference genes for steady state transcript quantification by qPCR in the diatom Phaeodactylum tricornutum with constitutive expression independent from time and light. Endocytobiosis Cell Res. J. Int. Soc. Endocytobiol. 2013, 24, 1–7. [Google Scholar]
- Chen, B.; Wan, C.; Mehmood, M.A.; Chang, J.S.; Bai, F.; Zhao, X. Manipulating environmental stresses and stress tolerance of microalgae for enhanced production of lipids and value-added products—A review. Bioresour. Technol. 2017, 244, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Sajjadi, B.; Chen, W.Y.; Raman, A.A.A.; Ibrahim, S. Microalgae lipid and biomass for biofuel production: A comprehensive review on lipid enhancement strategies and their effects on fatty acid composition. Renew. Sustain. Energy Rev. 2018, 97, 200–232. [Google Scholar] [CrossRef]
- Shi, K.; Gao, Z.; Shi, T.Q.; Song, P.; Ren, L.J.; Huang, H.; Ji, X.J. Reactive oxygen species-mediated cellular stress response and lipid accumulation in oleaginous microorganisms: The state of the art and future perspectives. Front. Microbiol. 2017, 8, 793. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, H.P.; Han, B.; Yu, X. Coupling of abiotic stresses and phytohormones for the production of lipids and high-value by-products by microalgae: A review. Bioresour. Technol. 2019, 274, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Villanova, V.; Fortunato, A.E.; Singh, D.; Bo, D.D.; Conte, M.; Obata, T.; Jouhet, J.; Fernie, A.R.; Marechal, E.; Falciatore, A.; et al. Investigating mixotrophic metabolism in the model diatom Phaeodactylum tricornutum. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.F.; Qin, Q.W.; Yan, S.K.; Huang, J.-L.; Liu, K.; Zhou, S.B. Biodiesel production from Chlorella vulgaris under nitrogen starvation in autotrophic, heterotrophic, and mixotrophic cultures. J. Appl. Phycol. 2019, 31, 1589–1596. [Google Scholar] [CrossRef]
- Kwak, H.S.; Kim, J.Y.H.; Woo, H.M.; Jin, E.S.; Min, B.K.; Sim, S.J. Synergistic effect of multiple stress conditions for improving microalgal lipid production. Algal Res. 2016, 19, 215–224. [Google Scholar] [CrossRef]
- Burch, A.R.; Franz, A.K. Combined nitrogen limitation and hydrogen peroxide treatment enhances neutral lipid accumulation in the marine diatom Phaeodactylum tricornutum. Bioresour. Technol. 2016, 219, 559–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Marchand, J.; Blanckaert, V.; Lukomska, E.; Ulmann, L.; Wielgosz-Collin, G.; Rabesaotra, V.; Moreau, B.; Bougaran, G.; Mimouni, V.; et al. Nitrogen and phosphorus limitations induce carbon partitioning and membrane lipid remodelling in the marine diatom Phaeodactylum tricornutum. Eur. J. Phycol. 2019, 54, 342–358. [Google Scholar] [CrossRef]
- Liang, J.; Wen, F.; Liu, J. Transcriptomic and lipidomic analysis of an EPA-containing Nannochloropsis sp. PJ12 in response to nitrogen deprivation. Sci. Rep. 2019, 9, 4540. [Google Scholar] [CrossRef] [Green Version]
- D’Ippolito, G.; Sardo, A.; Paris, D.; Vella, F.M.; Adelfi, M.G.; Botte, P.; Gallo, C.; Fontana, A. Potential of lipid metabolism in marine diatoms for biofuel production. Biotechnol. Biofuels 2015, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Nagappan, S.; Devendran, S.; Tsai, P.C.; Jayaraman, H.; Alagarsamy, V.; Pugazhendhi, A.; Ponnusamy, V.K. Metabolomics integrated with transcriptomics and proteomics: Evaluation of systems reaction to nitrogen deficiency stress in microalgae. Process Biochem. 2020, 91, 1–14. [Google Scholar] [CrossRef]
- Valenzuela, J.; Carlson, R.P.; Gerlach, R.; Cooksey, K.; Peyton, B.M.; Bothner, B.; Fields, M.W. Nutrient resupplementation arrests bio-oil accumulation in Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 2013, 97, 7049–7059. [Google Scholar] [CrossRef] [Green Version]
- Řezanka, T.; Lukavský, J.; Nedbalová, L.; Kolouchová, I.; Sigler, K. Effect of starvation on the distribution of positional isomers and enantiomers of triacylglycerol in the diatom Phaeodactylum tricornutum. Phytochemistry 2012, 80, 17–27. [Google Scholar] [CrossRef]
- Belhaj, N.; Arab-Tehrany, E.; Linder, M. Oxidative kinetics of salmon oil in bulk and in nanoemulsion stabilized by marine lecithin. Process Biochem. 2010, 45, 187–195. [Google Scholar] [CrossRef]
- Cui, Y.; Thomas-Hall, S.R.; Schenk, P.M. Phaeodactylum tricornutum microalgae as a rich source of omega-3 oil: Progress in lipid induction techniques towards industry adoption. Food Chem. 2019, 297, 124937. [Google Scholar] [CrossRef]
- Goiris, K.; Van Colen, W.; Wilches, I.; León-Tamariz, F.; De Cooman, L.; Muylaert, K. Impact of nutrient stress on antioxidant production in three species of microalgae. Algal Res. 2015, 7, 51–57. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Nitrogen starvation-induced cellular crosstalk of ROS-scavenging antioxidants and phytohormone enhanced the biofuel potential of green microalga Acutodesmus dimorphus. Biotechnol. Biofuels 2017, 10, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, A.; Malik, S.; Zhu, H.; Xu, J.; Nawaz, M.Z.; Nawaz, S.; Asraful Alam, M.; Mehmood, M.A. Cultivating microalgae in wastewater for biomass production, pollutant removal, and atmospheric carbon mitigation; a review. Sci. Total Environ. 2020, 704, 135303. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, C.; Mitra, M.; Bhayani, K.; Bharadwaj, S.V.V.; Ghosh, T.; Dubey, S.; Mishra, S. Abiotic stresses as tools for metabolites in microalgae. Bioresour. Technol. 2017, 244, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.R.; Senhorinho, G.N.A.; Scott, J.A. Microalgae under environmental stress as a source of antioxidants. Algal Res. 2020, 52, 102104. [Google Scholar] [CrossRef]
- Sun, X.M.; Ren, L.J.; Zhao, Q.Y.; Ji, X.J.; Huang, H. Microalgae for the production of lipid and carotenoids: A review with focus on stress regulation and adaptation. Biotechnol. Biofuels 2018, 11, 272. [Google Scholar] [CrossRef] [Green Version]
- Jeyakumar, B.; Asha, D.; Varalakshmi, P.; Kathiresan, S. Nitrogen repletion favors cellular metabolism and improves eicosapentaenoic acid production in the marine microalga Isochrysis sp. CASA CC 101. Algal Res. 2020, 47, 101877. [Google Scholar]
- Singh, P.; Baranwal, M.; Reddy, S.M. Antioxidant and cytotoxic activity of carotenes produced by Dunaliella salina under stress. Pharm. Biol. 2016, 54, 2269–2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patras, D.; Moraru, C.V.; Socaciu, C. Bioactive Ingredients from Microalgae: Food and Feed Applications. Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca. Food Sci. Technol. 2019, 76, 1. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef] [PubMed]
- Nordin, N.; Yusof, N.; Maeda, T.; Mustapha, N.A.; Mohd Yusoff, M.Z.; Raja Khairuddin, R.F. Mechanism of carbon partitioning towards starch and triacylglycerol in Chlorella vulgaris under nitrogen stress through whole-transcriptome analysis. Biomass Bioenergy 2020, 138, 105600. [Google Scholar] [CrossRef]
- Park, J.J.; Wang, H.; Gargouri, M.; Deshpande, R.R.; Skepper, J.N.; Holguin, F.O.; Juergens, M.T.; Shachar-Hill, Y.; Hicks, L.M.; Gang, D.R. The response of Chlamydomonas reinhardtii to nitrogen deprivation: A systems biology analysis. Plant J. 2015, 81, 611–624. [Google Scholar] [CrossRef]
- Bromke, M.A.; Giavalisco, P.; Willmitzer, L.; Hesse, H. Metabolic Analysis of Adaptation to Short-Term Changes in Culture Conditions of the Marine Diatom Thalassiosira pseudonana. PLoS ONE 2013, 8, e67340. [Google Scholar] [CrossRef]
- Corteggiani Carpinelli, E.; Telatin, A.; Vitulo, N.; Forcato, C.; D’Angelo, M.; Schiavon, R.; Vezzi, A.; Giacometti, G.M.; Morosinotto, T.; Valle, G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant 2014, 7, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Dou, X.; Wu, J.; Meng, F. Attenuation pathways of erythromycin and biochemical responses related to algal growth and lipid synthesis in a microalga-effluent system. Environ. Res. 2021, 195, 110873. [Google Scholar] [CrossRef]
- Huerlimann, R.; Heimann, K. Comprehensive guide to acetyl-carboxylases in algae. Crit. Rev. Biotechnol. 2013, 33, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Cheng, J.; Zhang, X.; Ye, Q.; Chen, S.; Zhou, J.; Cen, K. Transcriptome and key gene expression related to carbon metabolism and fatty acid synthesis of Chlorella vulgaris under a nitrogen starvation and phosphorus repletion regime. J. Appl. Phycol. 2019, 31, 2881–2893. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Zhan, J.; He, C.; Wang, Q. Comparative metabolic profiling of the lipid-producing green microalga Chlorella reveals that nitrogen and carbon metabolic pathways contribute to lipid metabolism. Biotechnol. Biofuels 2017, 1, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Cai, J.; Fei, X. Effect of the expression and knockdown of citrate synthase gene on carbon flux during triacylglycerol biosynthesis by green algae Chlamydomonas reinhardtii. BMC Biochem. 2013, 14, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, S.; Hérault, J.; Caruso, A.; Pencréac’h, G.; Come, M.; Gauvry, L.; Claverol, S.; Loiseau, C. Proteomics and expression studies on lipids and fatty acids metabolic genes in Isochrysis galbana under the combined influence of nitrogen starvation and sodium acetate supplementation. Bioresour. Technol. Rep. 2021, 15, 100714. [Google Scholar] [CrossRef]
- Remmers, I.M.; D’Adamo, S.; Martens, D.E.; de Vos, R.C.H.; Mumm, R.; America, A.H.P.; Cordewener, J.H.G.; Bakker, L.V.; Peters, S.A.; Wijffels, R.H.; et al. Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res. 2018, 35, 33–49. [Google Scholar] [CrossRef]
- Wan, M.; Liu, P.; Xia, J.; Rosenberg, J.N.; Oyler, G.A.; Betenbaugh, M.J.; Nie, Z.; Qiu, G. The effect of mixotrophy on microalgal growth, lipid content, and expression levels of three pathway genes in Chlorella sorokiniana. Appl. Microbiol. Biotechnol. 2011, 91, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Nagao, R.; Yokono, M.; Tomo, T.; Akimoto, S. Control mechanism of excitation energy transfer in a complex consisting of photosystem II and fucoxanthin chlorophyll a/c-binding protein. J. Phys. Chem. Lett. 2014, 5, 2983–2987. [Google Scholar] [CrossRef]
- Alipanah, L.; Rohloff, J.; Winge, P.; Bones, A.M.; Brembu, T. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 2015, 66, 6281–6296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abida, H.; Dolch, L.J.; Meï, C.; Villanova, V.; Conte, M.; Block, M.A.; Finazzi, G.; Bastien, O.; Tirichine, L.; Bowler, C.; et al. Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol. 2015, 167, 118–136. [Google Scholar] [CrossRef] [Green Version]









| Gene | Access Number | F/R Primer Sequence (5′–3′) | References |
|---|---|---|---|
| G6PDH | F. GCGAGAAATGGCACAAGG R. GTTCATCGCAGTCGGGAGA | [35] | |
| rbcL | MH064127.1 | F. CCAAGGTCCTGCTACTGGTG R. TCTCCAACGCATGAAGGGT | |
| FCP B | F. GCCGATATCCCCAATGGATTT R. CTTGGTCGAAGGAGTCCCATC | [36] | |
| ACCase, | F. GTTGCTTGACGCTGAACTGG R. CCTTCATGCGACCTGTCTTG | [37] | |
| Cit syn | F. TTATGAAGTCATGCCCGACA R. GGTCCCAGTACAGTTGCGAT | [37] | |
| Isocit DH | F. GGGCAGTCATGAAAGACGTT R. ATCCGTCAGCATATCACCGT | [37] | |
| RPS | F. AATTCCTCGAAGTCAACCAGG R. GTGCAAGAGACCGGACATAC | [38] | |
| TBP | F. ATCGATTTGTCAATCCACGAG R. ATACAGATTCTGTGTCCACGG | [38] |
| Fatty Acid | N+ | N− |
|---|---|---|
| 14:0 | 7.35 ± 0.54 a | 4.85 ± 0.61 b |
| 16:0 | 11.87 ± 0.72 a | 31.90 ± 1.57 b |
| 16:1n-7 | 31.12 ± 2.82 a | 43.35 ± 1.59 b |
| 16:2n-4 | 1.99 ± 0.62 a | 0.38 ± 0.02 b |
| 16:3n-4 | 12.12 ± 1.51 a | 1.07 ± 0.10 b |
| 18:1n-9 | 2.63 ± 1.59 | 4.90 ± 0.34 |
| 18:1n-7 | 1.13 ± 0.21 | 1.16 ± 0.18 |
| 18:2n-6 | 1.11 ± 0.20 a | 0.71 ± 0.11 b |
| 20:5n-3 EPA | 24.43 ± 1.85 a | 8.30 ± 0.80 b |
| 22:5n-3 | 1.88 ± 0.37 a | 0.69 ± 0.13 b |
| 22:6n-3 DHA | 1.62 ± 0.33 a | 0.71 ± 0.10 b |
| N+ | N− | |
|---|---|---|
| Polyene Index | 2.21 ± 0.32 a | 0.28 ± 0.04 b |
| n3/tot FA | 0.30 ± 0.04 a | 0.10 ± 0.01 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curcuraci, E.; Manuguerra, S.; Messina, C.M.; Arena, R.; Renda, G.; Ioannou, T.; Amato, V.; Hellio, C.; Barba, F.J.; Santulli, A. Culture Conditions Affect Antioxidant Production, Metabolism and Related Biomarkers of the Microalgae Phaeodactylum tricornutum. Antioxidants 2022, 11, 411. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11020411
Curcuraci E, Manuguerra S, Messina CM, Arena R, Renda G, Ioannou T, Amato V, Hellio C, Barba FJ, Santulli A. Culture Conditions Affect Antioxidant Production, Metabolism and Related Biomarkers of the Microalgae Phaeodactylum tricornutum. Antioxidants. 2022; 11(2):411. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11020411
Chicago/Turabian StyleCurcuraci, Eleonora, Simona Manuguerra, Concetta Maria Messina, Rosaria Arena, Giuseppe Renda, Theodora Ioannou, Vito Amato, Claire Hellio, Francisco J. Barba, and Andrea Santulli. 2022. "Culture Conditions Affect Antioxidant Production, Metabolism and Related Biomarkers of the Microalgae Phaeodactylum tricornutum" Antioxidants 11, no. 2: 411. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11020411