Rice Nudix Hydrolase OsNUDX2 Sanitizes Oxidized Nucleotides
Abstract
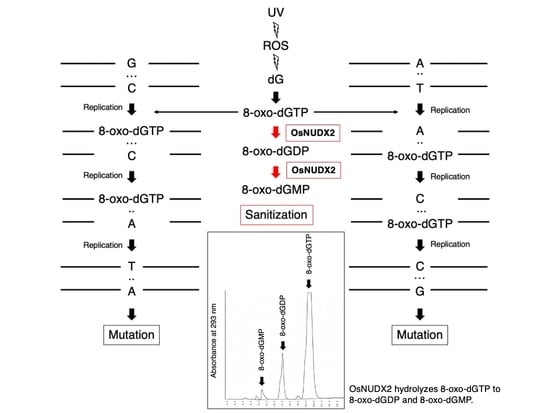
:1. Introduction
2. Materials and Methods
2.1. Plant Cultivation and UV Treatment
2.2. qRT-PCR Analysis
2.3. Expression and Purification of OsNUDX2
2.4. Enzyme and Protein Assays
2.5. Complementation Assay of Transcriptional Errors in mutT-Deficient E. coli
3. Results and Discussion
3.1. Classification of Proteins Encoded by Putative Rice NUDX Genes
3.2. Expression of Rice NUDX Genes under UV-C Irradiation
3.3. Purification and Enzymatic Characterization of Recombinant OsNUDX2
3.4. Prevention of Transcription Errors by OsNUDX2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danon, A.; Gallois, P. UV-C radiation induces apoptotic-like changes in Arabidopsis thaliana. FEBS Lett. 1998, 437, 131–136. [Google Scholar] [CrossRef]
- Jansen, M.A.K.; Gaba, V.; Greenberg, B.M. Higher plant and UV-B radiation: Balancing damage, repair and acclimation. Trends Plant Sci. 1998, 3, 131–135. [Google Scholar] [CrossRef]
- Zacchini, M.; de Agazio, M. Spread of oxidative damage and antioxidative response through cell layers of tobacco callus after UV-C treatment. Plant Physiol. Biochem. 2004, 42, 445–450. [Google Scholar] [CrossRef]
- Kasai, H.; Chung, M.H.; Yamamoto, F.; Ohtsuka, E.; Laval, J.; Grollman, A.P.; Nishimura, S. Formation, inhibition of formation, and repair of oxidative 8-hydroxyguanine DNA damage. Basic Life Sci. 1993, 61, 257–262. [Google Scholar]
- Ames, B.N.; Gold, L.S. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 1991, 250, 3–16. [Google Scholar] [CrossRef]
- Kasai, H.; Crain, P.F.; Kuchino, Y.; Nishimura, S.; Ootsuyama, A.; Tanooka, H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 1986, 7, 1849–1851. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Tsuzuki, T. Oxidative nucleotide damage: Consequences and prevention. Oncogene 2002, 21, 8895–8904. [Google Scholar] [CrossRef]
- Fowler, R.G.; White, S.J.; Koyama, C.; Moore, S.C.; Dunn, R.L.; Schaaper, R.M. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA Repair 2003, 2, 159–173. [Google Scholar] [CrossRef]
- Fujikawa, K.; Kamiya, H.; Yakushiji, H.; Nakabeppu, Y.; Kasai, H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001, 29, 449–454. [Google Scholar] [CrossRef]
- Furuichi, M.; Yoshida, M.C.; Oda, H.; Tajiri, T.; Nakabeppu, Y.; Tsuzuki, T.; Sekiguchi, M. Genomic structure and chromosome location of the human mutT homologue gene encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics 1994, 24, 485–490. [Google Scholar] [CrossRef]
- Ishibashi, T.; Hayakawa, H.; Sekiguchi, M. A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 2003, 4, 479–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishibashi, T.; Hayakawa, H.; Ito, R.; Miyazawa, M.; Yamagata, Y.; Sekiguchi, M. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res. 2005, 33, 3779–3784. [Google Scholar] [CrossRef] [PubMed]
- Maki, H.; Sekiguchi, M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 1992, 355, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y.; Sakumi, K.; Sakamoto, K.; Tsuchimoto, D.; Tsuzuki, T.; Nakatsu, Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol. Chem. 2006, 387, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Taddei, F.; Hayakawa, H.; Bouton, M.; Cirinesi, A.; Matic, I.; Sekiguchi, M.; Radman, M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 1997, 278, 128–130. [Google Scholar] [CrossRef]
- Bessman, M.J.; Frick, D.N.; O’Handley, S.F. The MutT proteins or “nudix” hydrolases, a family of versatile, widely distributed “house-cleaning” enzymes. J. Biol. Chem. 1996, 271, 25059–25062. [Google Scholar] [CrossRef]
- Kraszewska, E. The plant nudix hydrolase family. Acta Biochi. Pol. 2008, 55, 663–671. [Google Scholar] [CrossRef]
- McLennan, A.G. The nudix hydrolase superfamily. Cell Mol. Life Sci. 2006, 63, 123–142. [Google Scholar] [CrossRef]
- Xu, W.; Gauss, P.; Shen, J.Y.; Dunn, C.A.; Bessman, M.J. Three new nudix hydrolases from Escherichia coli. J. Biol. Chem. 2006, 281, 22794–22798. [Google Scholar] [CrossRef]
- Tanaka, S.; Kihara, M.; Sugimoto, M. Structure and molecular characterization of barley nudix hydrolase genes. Biosci. Biotechnol. Biochem. 2015, 79, 394–401. [Google Scholar] [CrossRef]
- Yoshimura, K.; Shigeoka, S. Versatile physiological functions of the nudix hydrolase family in arabidopsis. Biosci. Biotechnol. Biochem. 2015, 79, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Ueda, Y.; Yoshimura, K.; Shigeoka, S. Comprehensive analysis of cytosolic nudix hydrolases in Arabidopsis thaliana. J. Biol. Chem. 2005, 280, 25277–25283. [Google Scholar] [CrossRef]
- Yoshimura, K.; Ogawa, T.; Ueda, Y.; Shigeoka, S. AtNUDX1, an 8-oxo-7,8-dihydro-20-deoxyguanosine 5′-triphosphate pyrophosphohydrolase, is responsible for eliminating oxidized nucleotides in Arabidopsis. Plant Cell Physiol. 2007, 48, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Teranishi, M.; Ishida, H.; Kawasaki, J.; Takeuchi, A.; Yamaya, T.; Watanabe, M.; Makino, A.; Hidema, J. Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA. Plant J. 2011, 66, 433–442. [Google Scholar] [CrossRef]
- Du, H.; Liang, Y.; Pei, K.; Ma, K. UV radiation-responsive proteins in rice leaves: A proteomic analysis. Plant Cell Physiol. 2011, 52, 306–316. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, X.; Zhan, F.; Xie, C.; Zu, Y.; Li, Y.; Yue, M. Resistance-related physiological response of rice leaves to the compound stress of enhanced UV-B radiation and magnaporthe oryzae. J. Plant Interact. 2018, 13, 321–328. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nakanishi, H.; Takahashi, M.; Kawasaki, S.; Nishizawa, N.K.; Mori, S. In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2′-deoxymugineic acid to mugineic acid in transgenic rice. Planta 2001, 212, 864–871. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Millar, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972; pp. 352–355. [Google Scholar]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.; Iwamoto, M.; Abe, T.; et al. Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e1–e11. [Google Scholar] [CrossRef]
- Tompson, J.D.; Higgins, D.G.; Gibson, T.J. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Harwood, J.L.; Bach, T.J. A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 2012, 51, 95–148. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar] [PubMed]
- Henry, L.K.; Thomas, S.T.; Widhalm, J.R.; Lynch, J.H.; Davis, T.C.; Kessler, S.A.; Bohlmann, J.; Noel, J.P.; Dudareva, N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 2018, 4, 721–729. [Google Scholar] [CrossRef]





| Gene (Deduced Subfamily) | Locus ID | Position | Orthologue | (Subfamily) | Identity (%) |
|---|---|---|---|---|---|
| OsNUDX1 | Os06g0634300 | chr06:25718152..25722139 (−strand) | HvNUDX1 | (ADP ribose/NADH) | 82 |
| (ADP-ribose/NADH) | AtNUDX2 | (ADP ribose/NADH) | 57 | ||
| AtNUDX7 | (ADP ribose/NADH) | 47 | |||
| OsNUDX2 | Os02g0793300 | chr02:33710064..33719486 (+strand) | HvNUDX2 | 89 | |
| (DMAPP/IPP) | AtNUDX3 | (DMAPP/IPP) | 68 | ||
| OsNUDX3 | Os06g0255400 | chr06:8069204..8069983 (−strand) | HvNUDX3 | (ApnA/ppGpp) | 75 |
| (ApnA/ppGpp) | HvNUDX8 | (ApnA/ppGpp) | 61 | ||
| AtNUDX18 | 44 | ||||
| AtNUDX17 | 43 | ||||
| OsNUDX4 | Os05g0117500 | chr05:922037..926090 (+strand) | HvNUDX4 | (GDP mannose) | 74 |
| (GDP-mannose) | AtNUDX9 | (GDP mannose) | 59 | ||
| OsNUDX5 | Os05g0209400 | chr05:6766494..6768972 (+strand) | HvNUDX5 | (CoA) | 68 |
| (CoA) | HvNUDX14 | (CoA) | 49 | ||
| AtNUDX11 | (CoA) | 46 | |||
| AtNUDX15 | (CoA) | 46 | |||
| OsNUDX6 | Os04g0399300 | chr04:19735871..19739665 (+strand) | HvNUDX6 | (ApnA/ppGpp) | 58 |
| (ApnA/ppGpp) | AtNUDX12 | 43 | |||
| AtNUDX13 | (APnA/ppGpp) | 42 | |||
| OsNUDX7 | Os06g0129700 | chr06:1574614..1577176 (−strand) | HvNUDX7 | (ADP ribose/glucose) | 80 |
| (ADP-ribose/glucose) | AtNUDX14 | (ADP ribose/glucose) | 59 | ||
| OsNUDX8 | Os02g0734300 | chr02:30625668..30626865 (+strand) | HvNUDX8 | (ApnA) | 78 |
| (ApnA) | HvNUDX3 | (ApnA) | 66 | ||
| AtNUDX18 | 52 | ||||
| AtNUDX17 | 49 | ||||
| AtNUDX21 | 48 | ||||
| OsNUDX9 | Os06g0141166 | chr06:2157903..2160576 (−strand) | HvNUDX9 | (NADPH) | 79 |
| (NADPH) | AtNUDX19 | (NADPH) | 55 | ||
| OsNUDX10 | Os09g0322200 | chr09:9379558..9383831 (−strand) | HvNUDX10 | (Thiamine) | 79 |
| (Thiamine) | AtNUDX20 | (Thiamine) | 63 | ||
| AtNUDX24 | 60 | ||||
| OsNUDX11 | Os09g0553300 | chr09:21922941..21926057 (−strand) | HvNUDX11 | (FAD) | 72 |
| (FAD) | AtNUDX23 | (FAD) | 49 | ||
| OsNUDX12 | Os02g0520100 | chr02:18936409..18939517 (−strand) | HvNUDX6 | (ApnA/ppGpp) | 59 |
| (ApnA/ppGpp) | AtNUDX12 | 48 | |||
| AtNUDX13 | (ApnA/ppGpp) | 48 | |||
| OsNUDX13 | Os11g0531700 | chr11:19353108..19355768 (+strand) | HvNUDX6 | (ApnA/ppGpp) | 47 |
| (ApnA/ppGpp) | AtNUDX12 | 46 | |||
| AtNUDX13 | (ApnA/ppGpp) | 46 | |||
| OsNUDX14 | Os08g0375900 | chr08:17634789..17639334 (+strand) | HvNUDX14 | (CoA) | 70 |
| (CoA) | AtNUDX15 | (CoA) | 56 | ||
| AtNUDX11 | (CoA) | 53 | |||
| OsNUDX15 | Os07g0212300 | chr07:6137057..6141022 (−strand) | AtNUDX16 | (ApnA/ppGpp) | 62 |
| (ApnA/ppGpp) | AtNUDX13 | (ApnA/ppGpp) | 44 | ||
| AtNUDX12 | 42 | ||||
| OsNUDX16 | Os02g0805900 | chr02:34378712..34382107 (−strand) | AtDCP2 | (mRNA decap) | 58 |
| (mRNA decap) | |||||
| OsNUDX17 | Os06g0712200 | chr06:30117755..30118750 (+strand) | not identified |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, Y.; Rikiishi, K.; Sugimoto, M. Rice Nudix Hydrolase OsNUDX2 Sanitizes Oxidized Nucleotides. Antioxidants 2022, 11, 1805. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11091805
Kondo Y, Rikiishi K, Sugimoto M. Rice Nudix Hydrolase OsNUDX2 Sanitizes Oxidized Nucleotides. Antioxidants. 2022; 11(9):1805. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11091805
Chicago/Turabian StyleKondo, Yuki, Kazuhide Rikiishi, and Manabu Sugimoto. 2022. "Rice Nudix Hydrolase OsNUDX2 Sanitizes Oxidized Nucleotides" Antioxidants 11, no. 9: 1805. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11091805