Reduction of Oxidative Stress through Activating the Nrf2 mediated HO-1 Antioxidant Efficacy Signaling Pathway by MS15, an Antimicrobial Peptide from Bacillus velezensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. In Vitro Process of Isolation, Screening, and Culture Media Optimization
2.3. Optimizations of Culture Media Design by Response Surface Methodology and Statistical Analysis
2.4. Culture and Purification of Peptide MS15
2.5. Electrophoresis with Tricine SDS-PAGE and Bioassay (In Situ) Analysis
2.6. MALDI-TOF/MS for Molecular Weight Determination
2.7. pH and Thermal Stability of MS15
2.8. Evaluation of Antimicrobial Susceptibility
2.9. Bacterial Killing Kinetics Assay of Peptide MS15
2.10. DPPH (2,2-Diphenyl-1-Picrylhydrazyl) Radical Scavenging Capability Assay
2.11. ABTS (2,2′-Azino-bis (3-Ethylbenzthiazoline-6-Sulfonic Acid)) Radical Scavenging Capability Assay
2.12. Superoxide Radical Scavenging Capability Assay
2.13. FRAP (Ferric Reducing Antioxidant Power) Activity Assay
2.14. CUPRAC (Cupric Reducing Antioxidant Capacity) Activity Assay
2.15. ORAC (Oxygen Radicle Absorbance Capacity) Assay
2.16. Cellular NO (Nitric Oxide) Measurement in RAW 264.7 Cells
2.17. Intracellular ROS (Reactive Oxygen Species) Measurement in RAW 246.7 Cells
2.18. Cell Culture and Cell Cytotoxicity Assay
2.19. Western Blot Analysis with Cell Lysates
2.20. RT-PCR (Reverse Transcription-Polymerase Chain Reaction) Assay
2.21. Statistical Analysis
3. Results
3.1. Strain Isolation and Identification
3.2. Experimental Design and Box-Behnken Analysis by Response Surface Methodology
3.3. Culture Media, Purification, and Molecular Weight Resolve of MS15
3.4. Mass Spectroscopy Analysis by MALDI-TOF-MS
3.5. Stability Analysis of MS15
3.6. Assessment of Antimicrobial Susceptibility and Time-Kill Kinetics Analyses
3.7. Radical Scavenging Activity Assay
3.8. Reducing Power Measurement Assay
3.9. Cell Viability Assay
3.10. ROS Measurement and Nitric Oxide Inhibitory Assay
3.11. Effects of MS15 on Antioxidant Enzyme in RAW 264.7 Cells
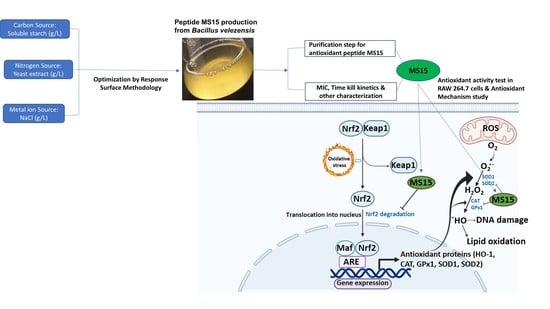
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.L.; Norhaizan, M.E.; Liew, W.-P.-P.; Rahman, H.S. Antioxidant and Oxidative Stress: A Mutual Interplay in Age-Related Diseases. Front. Pharmacol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahal, A.; Kumar, A.; Singh, V.; Yadav, B.; Tiwari, R.; Chakraborty, S.; Dhama, K. Oxidative Stress, Prooxidants, and Antioxidants: The Interplay. BioMed Res. Int. 2014, 2014, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa-Diez, C.; Miguel, V.; Mennerich, D.; Kietzmann, T.; Pérez, P.S.; Cadenas, S.; Lamas, S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Boil. 2015, 6, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative Stress and Neurodegenerative Diseases: A Review of Upstream and Downstream Antioxidant Therapeutic Options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Xing, L.; Fu, Q.; Zhou, G.; Zhang, W. A Review of Antioxidant Peptides Derived from Meat Muscle and By-Products. Antioxidants 2016, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Magesh, S.; Chen, Y.; Hu, L. Small Molecule Modulators of Keap1-Nrf2-ARE Pathway as Potential Preventive and Therapeutic Agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [Green Version]
- Velichkova, M.; Hasson, T. Keap1 Regulates the Oxidation-Sensitive Shuttling of Nrf2 into and out of the Nucleus via a Crm1-Dependent Nuclear Export Mechanism. Mol. Cell. Boil. 2005, 25, 4501–4513. [Google Scholar] [CrossRef] [Green Version]
- Fisgin, N.T.; Aydin, B.K.; Sarikaya, H.; Tanyel, E.; Esen, S.; Sunbul, M.; Leblebicioglu, H. Oxidative stress and antioxidant defense in patients with chronic hepatitis B. Clin. Lab. 2012, 58, 9–19. [Google Scholar]
- Daliri, E.B.-M.; Oh, D.-H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Konuray, G.; Erginkaya, Z. Potential Use of Bacillus coagulans in the Food Industry. Foods 2018, 7, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karami, Z.; Akbari-Adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Choi, Y.S.; Kim, Y.K.; Yoo, J.C. Immobilization of an alkaline endopolygalacturonase purified from Bacillus paralicheniformis exhibits bioscouring of cotton fabrics. Bioprocess Biosyst. Eng. 2018, 41, 1425–1436. [Google Scholar] [CrossRef]
- Lechevalier, H. A practical guide to generic identification of actinomycetes. In Bergey’s Man System Bacteria; Williams & Wilkins Company: Baltimore, MD, USA, 1989; pp. 2344–2347. [Google Scholar]
- Rahman, S.; Choi, Y.H.; Choi, Y.S.; Yoo, J.C. Glycin-rich antimicrobial peptide YD1 from B. amyloliquefaciens, induced morphological alteration in and showed affinity for plasmid DNA of E. coli. AMB Express 2017, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Kim, Y.K.; Cho, S.-S.; Jin, Y.-Y.; Suh, J.-W.; Lee, D.; Yoo, J.C. Response Surface Optimization of Culture Conditions for Cyclic Lipopeptide MS07 from Bacillus siamensis Reveals Diverse Insights Targeting Antimicrobial and Antibiofilm Activity. Processes 2020, 8, 744. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef]
- Regmi, S.; Choi, Y.S.; Choi, Y.H.; Kim, Y.K.; Cho, S.S.; Yoo, J.C.; Suh, J.-W. Antimicrobial peptide from Bacillus subtilis CSB138: Characterization, killing kinetics, and synergistic potency. Int. Microbiol. 2017, 20, 43–53. [Google Scholar] [PubMed]
- Hou, F.; Li, J.; Pan, P.; Xu, J.; Liu, L.; Liu, W.; Song, B.; Li, N.; Wan, J.; Gao, H. Isolation and characterisation of a new antimicrobial peptide from the skin of Xenopus laevis. Int. J. Antimicrob. Agents 2011, 38, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Qaoud, M.T.; Mohammed, G.K.; Abualhaijaa, A.; Knappe, D.; Hoffmann, R.; Al-Balas, Q.A. Antimicrobial and Antibiofilm Activity of UP-5, an Ultrashort Antimicrobial Peptide Designed Using Only Arginine and Biphenylalanine. Pharmaceuticals 2018, 11, 3. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Weir, M.D.; Fouad, A.F.; Xu, H.H.K. Time-kill behaviour against eight bacterial species and cytotoxicity of antibacterial monomers. J. Dent. 2013, 41, 881–891. [Google Scholar] [CrossRef] [Green Version]
- Nanjo, F.; Goto, K.; Seto, R.; Suzuki, M.; Sakai, M.; Hara, Y. Scavenging effects of tea catechins and their derivatives on 1,1-diphenyl-2-picrylhydrazyl radical. Free. Radic. Boil. Med. 1996, 21, 895–902. [Google Scholar] [CrossRef]
- Carrasco-Castilla, J.; Álvarez, A.J.H.; Jiménez-Martínez, C.; Jacinto-Hernández, C.; Alaiz, M.; Calle, J.G.; Vioque, J.; Dávila-Ortíz, G. Antioxidant and metal chelating activities of peptide fractions from phaseolin and bean protein hydrolysates. Food Chem. 2012, 135, 1789–1795. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Alam, B.; Quan, K.T.; Kwon, K.-R.; Ju, M.-K.; Choi, H.-J.; Lee, J.S.; Yoon, J.-I.; Majumder, R.; Rather, I.A.; et al. Antioxidant efficacy and the upregulation of Nrf2-mediated HO-1 expression by (+)-lariciresinol, a lignan isolated from Rubia philippinensis, through the activation of p38. Sci. Rep. 2017, 7, 46035. [Google Scholar] [CrossRef] [Green Version]
- Benzie, I.F.; Strain, J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Apak, R.; Güçlü, K.; Özyürek, M.; Karademir, S.E. Novel Total Antioxidant Capacity Index for Dietary Polyphenols and Vitamins C and E, Using Their Cupric Ion Reducing Capability in the Presence of Neocuproine: CUPRAC Method. J. Agric. Food Chem. 2004, 52, 7970–7981. [Google Scholar] [CrossRef]
- Ou, B.; Huang, D.; Hampsch-Woodill, M.; Flanagan, J.A.; Deemer, E.K. Analysis of Antioxidant Activities of Common Vegetables Employing Oxygen Radical Absorbance Capacity (ORAC) and Ferric Reducing Antioxidant Power (FRAP) Assays: A Comparative Study. J. Agric. Food Chem. 2002, 50, 3122–3128. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Choi, Y.H.; Choi, Y.S.; Alam, B.; Lee, S.-H.; Yoo, J.C. A novel antioxidant peptide, purified from Bacillus amyloliquefaciens, showed strong antioxidant potential via Nrf-2 mediated heme oxygenase-1 expression. Food Chem. 2018, 239, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Bender, C.; Graziano, S.; Zimmerman, B.F.; Weidlich, H.H. Antioxidant potential of aqueous plant extracts assessed by the cellular antioxidant activity assay. Am. J. Biol. Life Sci. 2014, 2, 72–79. [Google Scholar]
- Choi, Y.H.; Cho, S.S.; Simkhada, J.R.; Rahman, S.; Choi, Y.S.; Kim, C.S.; Yoo, J.C. A novel multifunctional peptide oligomer of bacitracin with possible bioindustrial and therapeutic applications from a Korean food-source Bacillus strain. PLoS ONE 2017, 12, e0176971. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Cha, H.-J.; Choi, E.O.; Kim, C.H.; Kim, G.-Y.; Yoo, Y.H.; Hwang, H.-J.; Park, H.T.; Yoon, H.M.; Choi, Y.H. Activation of the Nrf2/HO-1 signaling pathway contributes to the protective effects of baicalein against oxidative stress-induced DNA damage and apoptosis in HEI193 Schwann cells. Int. J. Med. Sci. 2019, 16, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regmi, S.; Choi, Y.H.; Choi, Y.S.; Kim, M.R.; Yoo, J.C. Antimicrobial peptide isolated from Bacillus amyloliquefaciens K14 revitalizes its use in combinatorial drug therapy. Folia Microbiol. 2016, 62, 127–138. [Google Scholar] [CrossRef]
- Van Der Weide, H.; Brunetti, J.; Pini, A.; Bracci, L.; Ambrosini, C.; Lupetti, P.; Paccagnini, E.; Gentile, M.; Bernini, A.; Niccolai, N.; et al. Investigations into the killing activity of an antimicrobial peptide active against extensively antibiotic-resistant K. pneumon iae and P. aeruginosa. Biochim. Biophys. Acta (BBA) Biomembr. 2017, 1859, 1796–1804. [Google Scholar] [CrossRef]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of action of antimicrobial peptide P5 truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB Express 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A. Cellular Defenses against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef] [Green Version]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef] [Green Version]
- Lü, J.-M.; Lin, P.H.; Yao, Q.; Chen, C. Chemical and molecular mechanisms of antioxidants: Experimental approaches and model systems. J. Cell. Mol. Med. 2009, 14, 840–860. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Gao, J.; Xue, Z.; Zhang, Z.; Wang, H.; Wang, X. Purification and identification of antioxidant peptides from chickpea (Cicer arietinum L.) albumin hydrolysates. LWT Food Sci. Technol. 2013, 50, 591–598. [Google Scholar] [CrossRef]
- Luisi, G.; Stefanucci, A.; Zengin, G.; Dimmito, M.P.; Mollica, A. Anti-Oxidant and Tyrosinase Inhibitory In Vitro Activity of Amino Acids and Small Peptides: New Hints for the Multifaceted Treatment of Neurologic and Metabolic Disfunctions. Antioxidants 2018, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-K.; Jang, H.-D. Nrf2-Mediated HO-1 Induction Coupled with the ERK Signaling Pathway Contributes to Indirect Antioxidant Capacity of Caffeic Acid Phenethyl Ester in HepG2 Cells. Int. J. Mol. Sci. 2014, 15, 12149–12165. [Google Scholar] [CrossRef] [Green Version]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free. Radic. Boil. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Wu, J.Q.; Kosten, T.R.; Zhang, X.Y. Free radicals, antioxidant defense systems, and schizophrenia. Prog. Neuro-Psychopharmacol. Boil. Psychiatry 2013, 46, 200–206. [Google Scholar] [CrossRef]
- Zou, X.; Gao, J.; Zheng, Y.; Wang, X.; Chen, C.; Cao, K.; Xu, J.; Li, Y.; Lu, W.; Liu, J.; et al. Zeaxanthin induces Nrf2-mediated phase II enzymes in protection of cell death. Cell Death Dis. 2014, 5, e1218. [Google Scholar] [CrossRef] [Green Version]
- Chun, K.; Alam, B.; Son, H.-U.; Lee, S.-H. Effect of Novel Compound LX519290, a Derivative of l-allo Threonine, on Antioxidant Potential in Vitro and in Vivo. Int. J. Mol. Sci. 2016, 17, 1451. [Google Scholar] [CrossRef]
- Hassmann, U.; Volz, N.; Teller, N.; Haupt, L.M.; Bakuradze, T.; Eisenbrand, G.; Bytof, G.; Lantz, I.; Griffiths, L.R.; Marko, D. Induction of antioxidative Nrf2 gene transcription by coffee in humans: Depending on genotype? Mol. Boil. Rep. 2012, 39, 7155–7162. [Google Scholar] [CrossRef] [Green Version]







| Code | Variables of Choice (Independent) | Up Level | Midpoint | Down Level | Generated Optimum Factor |
|---|---|---|---|---|---|
| (+1) | (0) | (−1) | |||
| A | Soluble starch (g/L) | 25 | 15 | 5 | 13.42 |
| B | Yeast extract (g/L) | 15 | 10 | 5 | 7.64 |
| C | NaCl (g/L) | 0.15 | 0.1 | 0.05 | 0.089 |
| Run | Soluble Starch (g/L) | Yeast Extract (g/L) | NaCl (g/L) | Bacteriocin Activity of MS15 (AU/mL) | |
|---|---|---|---|---|---|
| A | B | C | Actual Value x | Predicted Value | |
| 1 | 15 | 10 | 0.1 | 12,138.00 | 12,205.80 |
| 2 | 5 | 15 | 0.1 | 9460.00 | 9166.00 |
| 3 | 15 | 10 | 0.1 | 11,940.00 | 12,205.80 |
| 4 | 5 | 10 | 0.05 | 8650.00 | 8782.25 |
| 5 | 15 | 5 | 0.05 | 10,450.00 | 10,072.25 |
| 6 | 5 | 10 | 0.15 | 10,113.00 | 10,029.25 |
| 7 | 25 | 10 | 0.05 | 11,146.00 | 11,229.75 |
| 8 | 15 | 10 | 0.1 | 12,268.00 | 12,205.80 |
| 9 | 15 | 5 | 0.15 | 11,329.00 | 11,167.25 |
| 10 | 25 | 5 | 0.1 | 9336.00 | 9630.00 |
| 11 | 5 | 5 | 0.1 | 7622.00 | 7867.50 |
| 12 | 15 | 10 | 0.1 | 12,316.00 | 12,205.80 |
| 13 | 25 | 15 | 0.1 | 11,560.00 | 11,314.50 |
| 14 | 15 | 15 | 0.05 | 11,742.00 | 11,903.75 |
| 15 | 15 | 15 | 0.15 | 11,941.00 | 12,318.75 |
| 16 | 25 | 10 | 0.15 | 11,625.00 | 11,492.75 |
| 17 | 15 | 10 | 0.1 | 12,367.00 | 12,205.80 |
| Purification Steps | Vol (mL) | Total Protein (mg) | Total Activity (AU) | Specific Activity (AU/mg) | Purification Fold | Recovery (%) |
|---|---|---|---|---|---|---|
| Cell free crude sample | 935 | 388.5 | 270,000 | 694.98 | 1 | 100 |
| Ammonium sulphate pallet aliquots | 52 | 98.45 | 156,000 | 1584.56 | 2.28 | 57.78 |
| Sephadex G-50 gel | 8 | 7.23 | 38,000 | 5255.88 | 7.56 | 14.07 |
| DEAE Sephadex A-50 gel | 2 | 0.94 | 16,000 | 17,021.27 | 24.49 | 5.92 |
| Microorganisms | MIC of MS15 (µg/mL) | MBC of MS15 (µg/mL) | ||
|---|---|---|---|---|
| MS15 | Bacitracin | Vancomycin | ||
| Gram-negative bacteria | ||||
| Escherichia coli KCTC 1923 | 40 | 160 | 80 | 80 |
| Pseudomonas aeruginosa KCTC 1637 | 160 | >160 | >160 | 320 |
| Salmonella typhimurium KCTC 1925 | 40 | 80 | 40 | 80 |
| Alcaligenes faecalis ATCC 1004 | 160 | 160 | 80 | 320 |
| Extended-spectrum beta-lactamase V4 (Escherichia coli) | 80 | 160 | 80 | 80 |
| Extended-spectrum beta-lactamase W1 | 40 | 80 | 40 | 80 |
| Extended-spectrum beta-lactamase 31 | 40 | 80 | 40 | 80 |
| Gram-positive bacteria | ||||
| Vancomycin-resistant Enterococci 4 | 160 | 80 | 160 | 320 |
| Vancomycin-resistant Enterococci 89 | 80 | >160 | >160 | 160 |
| Vancomycin-resistant Enterococci 98 | 80 | >160 | >160 | 160 |
| Staphylococcus aureus KCTC 1928 | 160 | >160 | >160 | 160 |
| Methicillin-resistant Staphylococcus aureus 5-3 | 2.5 | 5 | 2.5 | 5 |
| Methicillin-resistant Staphylococcus aureus B15 | 40 | 160 | 80 | 120 |
| Mycobacterium smegmatis ATCC 9341 | 20 | 40 | 2.5 | 40 |
| Micrococcus luteus ATCC 9341 | 40 | 40 | 2.5 | 40 |
| Enterococcus faecalis ATCC 29212 | 20 | 5 | 2.5 | 20 |
| Bacillus subtilis ATCC 6633 | 10 | 20 | 0.8 | 20 |
| Vancomycin-resistant Staphylococcus aureus | 80 | 160 | >160 | 160 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.M.; Kim, Y.K.; Bilkis, T.; Suh, J.-W.; Lee, D.Y.; Yoo, J.C. Reduction of Oxidative Stress through Activating the Nrf2 mediated HO-1 Antioxidant Efficacy Signaling Pathway by MS15, an Antimicrobial Peptide from Bacillus velezensis. Antioxidants 2020, 9, 934. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9100934
Khan MM, Kim YK, Bilkis T, Suh J-W, Lee DY, Yoo JC. Reduction of Oxidative Stress through Activating the Nrf2 mediated HO-1 Antioxidant Efficacy Signaling Pathway by MS15, an Antimicrobial Peptide from Bacillus velezensis. Antioxidants. 2020; 9(10):934. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9100934
Chicago/Turabian StyleKhan, Md Maruf, Young Kyun Kim, Tahmina Bilkis, Joo-Won Suh, Dae Young Lee, and Jin Cheol Yoo. 2020. "Reduction of Oxidative Stress through Activating the Nrf2 mediated HO-1 Antioxidant Efficacy Signaling Pathway by MS15, an Antimicrobial Peptide from Bacillus velezensis" Antioxidants 9, no. 10: 934. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9100934