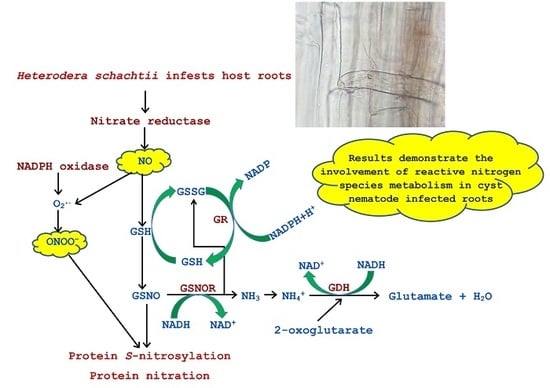
Cyst Nematode Infection Elicits Alteration in the Level of Reactive Nitrogen Species, Protein S-Nitrosylation and Nitration, and Nitrosoglutathione Reductase in Arabidopsis thaliana Roots
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Condition and Cyst Nematode Inoculation
2.2. Detection of Reactive Nitrogen Species with Confocal Laser Scanning Microscopy
2.3. Crude Extract Preparation
2.4. Profiling of S-Nitrosylated Proteins Patterns
2.5. Profiling of 3-Nitrotyrosine-Containing Proteins Patterns
2.6. Glutathione Reductase (GR) Activity
2.7. Glutathione Peroxidase (GPx) Activity
2.8. Nitrosoglutathione Reductase (GSNOR): Enzymatic Activity and Protein Level
2.8.1. Enzymatic Activity
2.8.2. Protein Gel Blot Analysis
2.8.3. Transmission Electron Microscopy (TEM)
2.8.4. Profiling of GSNOR Activity by Zymography in Native Polyacrylamide Gel
2.9. RNA Isolation and cDNA Synthesis
2.10. Quantitative Real-Time PCR
2.11. Statistics
3. Results
3.1. Generation and Localization of Reactive Nitrogen Species
3.2. Protein S-Nitrosylation and Nitration
3.3. GSNOR Protein Level and Enzyme Activity
3.4. Activity of Glutathione-Dependent Enzymes
3.5. Alterations in Gene Transcript Level of GSNOR and HB1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turkan, I. ROS and RNS: Key signalling molecules in plants. J. Exp. Bot. 2018, 69, 3313–3315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floryszak-Wieczorek, J.; Arasimowicz, M.; Milczarek, G.; Jelen, H.; Jackowiak, H. Only an early nitric oxide burst and the following wave of secondary nitric oxide generation enhanced effective defence responses of pelargonium to a necrotrophic pathogen. New Phytol. 2007, 175, 718–730. [Google Scholar] [CrossRef]
- Mai, V.C.; Drzewiecka, K.; Jeleń, H.; Narożna, D.; Rucińska-Sobkowiak, R.; Kęsy, J.; Floryszak-Wieczorek, J.; Gabryś, B.; Morkunas, I. Differential induction of Pisum sativum defense signaling molecules in response to pea aphid infestation. Plant Sci. 2014, 221, 1–12. [Google Scholar] [CrossRef]
- Morkunas, I.; Formela, M.; Floryszak-Wieczorek, J.; Marczak, Ł.; Narożna, D.; Nowak, W.; Bednarski, W. Cross-talk interactions of exogenous nitric oxide and sucrose modulates phenylpropanoid metabolism in yellow lupine embryo axes infected with Fusarium oxysporum. Plant Sci. 2013, 211, 102–121. [Google Scholar] [CrossRef] [Green Version]
- Nawrocka, J.; Gromek, A.; Małolepsza, U. Nitric oxide as a beneficial signaling molecule in Trichoderma atroviride TRS25-induced systemic defense responses of cucumber plants against Rhizoctonia solani. Front. Plant Sci. 2019, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Wojtaszek, P. Nitric oxide in plants: To NO or not to NO. Phytochemistry 2000, 54, 1–4. [Google Scholar] [CrossRef]
- Yamasaki, H. Nitrite-dependent nitric oxide production pathway: Implications for involvement of active nitrogen species in photoinhibition in vivo. Philos. Trans. R. Soc. Biol. Sci. 2000, 355, 1477–1488. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.J.; Igamberdiev, A.U.; Manjunatha, G.; Segu, S.; Moran, J.F.; Neelawarne, B.; Bauwe, H.; Kaiser, W.M. The emerging roles of nitric oxide (NO) in plant mitochondria. Plant Sci. 2011, 181, 520–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, K.; Majola, A.; Gokul, A.; Keyster, M.; Ludidi, N.; Egbichi, I. Inhibition of NOS-like activity in maize alters the expression of genes involved in H2O2 scavenging and glycine betaine biosynthesis. Sci. Rep. 2018, 8, 12628. [Google Scholar] [CrossRef] [PubMed]
- Arasimowicz-Jelonek, M.; Floryszak-Wieczorek, J.; Izbiańska, K.; Gzyl, J.; Jelonek, T. Implication of peroxynitrite in defence responses of potato to Phytophthora Infestans. Plant Pathol. 2016, 65, 754–766. [Google Scholar] [CrossRef] [Green Version]
- Vandelle, E.; Delledonne, M. Peroxynitrite formation and function in plants. Plant Sci. 2011, 181, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Friso, G.; van Wijk, K.J. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Nitro-oxidative stress vs. oxidative or nitrosative stress in higher plants. New Phytol. 2013, 199, 633–635. [Google Scholar] [CrossRef]
- Jahnová, J.; Luhová, L.; Petřivalský, M. S-nitrosoglutathione reductase-the master regulator of protein S-nitrosation in plant NO signaling. Plants 2019, 8, 48. [Google Scholar] [CrossRef] [Green Version]
- Hess, D.T.; Stamler, J.S. Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 2012, 287, 4411–4418. [Google Scholar] [CrossRef] [Green Version]
- Labudda, M. Ascorbate-glutathione pathway as an important player in redox regulation in nematode-infested plants: What we have learned so far. Physiol. Mol. Plant Pathol. 2018, 103, 47–53. [Google Scholar] [CrossRef]
- Perazzolli, M.; Dominici, P.; Romero-Puertas, M.C.; Zago, E.; Zeier, J.; Sonoda, M.; Lamb, C.; Delledonne, M. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 2004, 16, 2785–2794. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Cieśla, J.; Sobczak, M.; Dzik, J.M. Arginase activity in Arabidopsis thaliana infected with Heterodera schachtii. Plant Pathol. 2016, 65, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Siddique, S.; Matera, C.; Radakovic, Z.S.; Hasan, M.S.; Gutbrod, P.; Różańska, E.; Sobczak, M.; Torres, M.A.; Grundler, F.M.W. Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci. Signal. 2014, 7, ra33. [Google Scholar] [CrossRef] [Green Version]
- Melillo, M.T.; Leonetti, P.; Leone, A.; Veronico, P.; Bleve-Zacheo, T. ROS and NO production in compatible and incompatible tomato-Meloidogyne incognita interactions. Eur. J. Plant Pathol. 2011, 130, 489–502. [Google Scholar] [CrossRef]
- Zhou, J.; Jia, F.; Shao, S.; Zhang, H.; Li, G.; Xia, X.; Zhou, Y.; Yu, J.; Shi, K. Involvement of nitric oxide in the jasmonate-dependent basal defense against root-knot nematode in tomato plants. Front. Plant Sci. 2015, 6, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labudda, M.; Różańska, E.; Muszyńska, E.; Marecka, D.; Głowienka, M.; Roliński, P.; Prabucka, B. Heterodera schachtii infection affects nitrogen metabolism in Arabidopsis thaliana. Plant Pathol. 2020, 69, 794–803. [Google Scholar] [CrossRef]
- Gietler, M.; Nykiel, M.; Orzechowski, S.; Fettke, J.; Zagdańska, B. Proteomic analysis of S-nitrosylated and S-glutathionylated proteins in wheat seedlings with different dehydration tolerances. Plant Physiol. Biochem. 2016, 108, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Labudda, M.; Różańska, E.; Czarnocka, W.; Sobczak, M.; Dzik, J.M. Systemic changes in photosynthesis and reactive oxygen species homeostasis in shoots of Arabidopsis thaliana infected with the beet cyst nematode Heterodera schachtii. Mol. Plant Pathol. 2018, 19, 1690–1704. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, A.; Ueda, M.; Morikawa, H. Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 2002, 515, 20–24. [Google Scholar] [CrossRef] [Green Version]
- Golinowski, W.; Grundler, F.M.W.; Sobczak, M. Changes in the structure of Arabidopsis thaliana during female development of the plant-parasitic nematode Heterodera schachtii. Protoplasma 1996, 194, 103–116. [Google Scholar] [CrossRef]
- Brorson, S.H. Comparison of the immunolabeling of heated and deplasticized epoxy sections on the electron microscopical level. Micron 1999, 30, 319–324. [Google Scholar] [CrossRef]
- Kubienová, L.; Tichá, T.; Luhová, L.; Petřivalský, M. Detection of S-nitrosoglutathione reductase activity in plants. In Plant Nitric Oxide: Methods and Protocols; Gupta, K., Ed.; Springer: New York, NY, USA, 2016; pp. 175–189. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Prabucka, B.; Muszyńska, E.; Marecka, D.; Kozak, M.; Dababat, A.A.; Sobczak, M. Activity profiling of barley vacuolar processing enzymes provides new insights into the plant and cyst nematode interaction. Mol. Plant Pathol. 2020, 21, 38–52. [Google Scholar] [CrossRef] [Green Version]
- Hancock, J.T.; Desikan, R.; Clarke, A.; Hurst, R.D.; Neill, S.J. Cell signalling following plant/pathogen interactions involves the generation of reactive oxygen and reactive nitrogen species. Plant Physiol. Biochem. 2002, 40, 611–617. [Google Scholar] [CrossRef]
- Sidonskaya, E.; Schweighofer, A.; Shubchynskyy, V.; Kammerhofer, N.; Hofmann, J.; Wieczorek, K.; Meskiene, I. Plant resistance against the parasitic nematode Heterodera schachtii is mediated by MPK3 and MPK6 kinases, which are controlled by the MAPK phosphatase AP2C1 in Arabidopsis. J. Exp. Bot. 2016, 67, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeece, B.T.; Sharma, K.; Lawrence, G.W.; Lawrence, K.S.; Klink, V.P. The mitogen activated protein kinase (MAPK) gene family functions as a cohort during the Glycine max defense response to Heterodera glycines. Plant Physiol. Biochem. 2019, 137, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Du, Y.; Li, Y.; Ren, D.; Song, C.P. Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 2010, 22, 2981–2998. [Google Scholar] [CrossRef] [Green Version]
- Siddique, S.; Endres, S.; Sobczak, M.; Radakovic, Z.S.; Fragner, L.; Grundler, F.M.W.; Weckwerth, W.; Tenhaken, R.; Bohlmann, H. Myo-inositol oxygenase is important for the removal of excess myo-inositol from syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytol. 2014, 201, 476–485. [Google Scholar] [CrossRef] [Green Version]
- Mata-Pérez, C.; Begara-Morales, J.C.; Chaki, M.; Sánchez-Calvo, B.; Valderrama, R.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Protein tyrosine nitration during development and abiotic stress response in plants. Front. Plant Sci. 2016, 7, 1699. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; Padilla, M.N.; Barroso, J.B. The function of S-nitrosothiols during abiotic stress in plants. J. Exp. Bot. 2019, 70, 4429–4439. [Google Scholar] [CrossRef]
- Gaut, J.P.; Byun, J.; Tran, H.D.; Lauber, W.M.; Carroll, J.A.; Hotchkiss, R.S.; Belaaouaj, A.; Heinecke, J.W. Myeloperoxidase produces nitrating oxidants in vivo. J. Clin. Investig. 2002, 109, 1311–1319. [Google Scholar] [CrossRef]
- Sakihama, Y.; Tamaki, R.; Shimoji, H.; Ichiba, T.; Fukushi, Y.; Tahara, S.; Yamasaki, H. Enzymatic nitration of phytophenolics: Evidence for peroxynitrite-independent nitration of plant secondary metabolites. FEBS Lett. 2003, 553, 377–380. [Google Scholar] [CrossRef] [Green Version]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 2018, 71, 142–154. [Google Scholar] [CrossRef] [Green Version]
- Demarchi, F.; Bertoli, C.; Copetti, T.; Tanida, I.; Brancolini, C.; Eskelinen, E.L.; Schneider, C. Calpain is required for macroautophagy in mammalian cells. J. Cell Biol. 2006, 175, 595–605. [Google Scholar] [CrossRef] [Green Version]
- Labudda, M.; Różańska, E.; Szewińska, J.; Sobczak, M.; Dzik, J.M. Protease activity and phytocystatin expression in Arabidopsis thaliana upon Heterodera schachtii infection. Plant Physiol. Biochem. 2016, 109, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Yun, B.W.; Kang, J.G.; Raja, M.U.; Kwon, E.; Sorhagen, K.; Chu, C.; Wang, Y.; Loake, G.J. Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 2008, 59, 147–154. [Google Scholar] [CrossRef] [PubMed]










© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labudda, M.; Różańska, E.; Gietler, M.; Fidler, J.; Muszyńska, E.; Prabucka, B.; Morkunas, I. Cyst Nematode Infection Elicits Alteration in the Level of Reactive Nitrogen Species, Protein S-Nitrosylation and Nitration, and Nitrosoglutathione Reductase in Arabidopsis thaliana Roots. Antioxidants 2020, 9, 795. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9090795
Labudda M, Różańska E, Gietler M, Fidler J, Muszyńska E, Prabucka B, Morkunas I. Cyst Nematode Infection Elicits Alteration in the Level of Reactive Nitrogen Species, Protein S-Nitrosylation and Nitration, and Nitrosoglutathione Reductase in Arabidopsis thaliana Roots. Antioxidants. 2020; 9(9):795. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9090795
Chicago/Turabian StyleLabudda, Mateusz, Elżbieta Różańska, Marta Gietler, Justyna Fidler, Ewa Muszyńska, Beata Prabucka, and Iwona Morkunas. 2020. "Cyst Nematode Infection Elicits Alteration in the Level of Reactive Nitrogen Species, Protein S-Nitrosylation and Nitration, and Nitrosoglutathione Reductase in Arabidopsis thaliana Roots" Antioxidants 9, no. 9: 795. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9090795