Tauroursodeoxycholic Acid (TUDCA)—Lipid Interactions and Antioxidant Properties of TUDCA Studied in Model of Photoreceptor Membranes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Lipids and Spin Labels
2.2. Preparation of Liposomes
2.3. Singlet Oxygen Quenching Measurements
2.4. EPR Oximetry
2.5. EPR Measurement of Structural Properties of Lipid Bilayers
2.6. Molecular Dynamics Simulation
2.7. Isolation of Detergent–Resistant (DRM) and Detergent–Soluble (DSM) Membrane Fractions
2.8. Raman Spectroscopy (RS)
3. Results and Discussions
3.1. Interaction of TUDCA with Singlet Oxygen
3.2. Reactivity of TUDCA towards Free Radicals
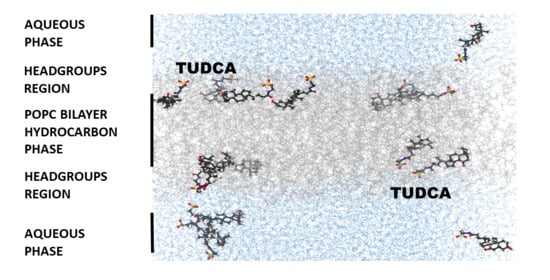
3.3. Location of TUDCA in the Membrane—Molecular Dynamics Simulation
3.4. Distribution of TUDCA between DRM and DSM Domains
3.5. Effect of TUDCA on Membrane Structural Properties
3.5.1. Polarity Profiles
3.5.2. Order Parameter
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hanafi, N.I.; Mohamed, A.S.; Sheikh Abdul Kadir, S.H.; Othman, M.H.D. Overview of Bile Acids Signaling and Perspective on the Signal of Ursodeoxycholic Acid, the Most Hydrophilic Bile Acid, in the Heart. Biomolecules 2018, 8, 159. [Google Scholar] [CrossRef] [Green Version]
- Berger, E.; Haller, D. Structure-function analysis of the tertiary bile acid TUDCA for the resolution of endoplasmic reticulum stress in intestinal epithelial cells. Biochem. Biophys. Res. Commun. 2011, 409, 610–615. [Google Scholar] [CrossRef]
- Hagey, L.R.; Crombie, D.L.; Espinosa, E.; Carey, M.C.; Igimi, H.; Hofmann, A.F. Ursodeoxycholic acid in the Ursidae: Biliary bile acids of bears, pandas, and related carnivores. J. Lipid Res. 1993, 34, 1911–1917. [Google Scholar] [CrossRef]
- Solá, S.; Garshelis, D.L.; Amaral, J.D.; Noyce, K.V.; Coy, P.L.; Steer, C.J.; Iaizzo, P.A.; Rodrigues, C.M.P. Plasma levels of ursodeoxycholic acid in black bears, Ursus americanus: Seasonal changes. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2006, 143, 204–208. [Google Scholar] [CrossRef]
- Vang, S.; Longley, K.; Steer, C.J.; Low, W.C. The Unexpected Uses of Urso- and Tauroursodeoxycholic Acid in the Treatment of Non-liver Diseases. Glob. Adv. Health Med. 2014, 3, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Ramalho, R.M.; Viana, R.J.; Low, W.C.; Steer, C.J.; Rodrigues, C.M. Bile acids and apoptosis modulation: An emerging role in experimental Alzheimer’s disease. Trend. Mol. Med. 2008, 14, 54–62. [Google Scholar] [CrossRef]
- Keene, C.D.; Rodrigues, C.M.; Eich, T.; Chhabra, M.S.; Steer, C.J.; Low, W.C. Tauroursodeoxycholic acid, a bile acid, is neuroprotective in a transgenic animal model of Huntington’s disease. Proc. Natl. Acad. Sci. USA 2002, 99, 10671–10676. [Google Scholar] [CrossRef] [Green Version]
- Grant, S.M.; DeMorrow, S. Bile Acid Signaling in Neurodegenerative and Neurological Disorders. Int. J. Mol. Sci. 2020, 21, 5982. [Google Scholar] [CrossRef]
- Boatright, J.H.; Moring, A.G.; McElroy, C.; Phillips, M.J.; Do, V.T.; Chang, B.; Hawes, N.L.; Boyd, A.P.; Sidney, S.S.; Stewart, R.E.; et al. Tool from ancient pharmacopoeia prevents vision loss. Mol. Vis. 2006, 12, 1706–1714. [Google Scholar]
- Feng, Y.; Siu, K.; Wang, N.; Ng, K.-M.; Tsao, S.-W.; Nagamatsu, T.; Tong, Y. Bear bile: Dilemma of traditional medicinal use and animal protection. J. Ethnobiol. Ethnomed. 2009, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Ludolph, A.C. The TUDCA trial—Innovative trial designs for amyotrophic lateral sclerosis drugs? Eur. J. Neurol. 2016, 23, 11–12. [Google Scholar] [CrossRef]
- Castro-Caldas, M.; Carvalho, A.N.; Rodrigues, E.; Henderson, C.J.; Wolf, C.R.; Rodrigues, C.M.; Gama, M.J. Tauroursodeoxycholic acid prevents MPTP-induced dopaminergic cell death in a mouse model of Parkinson’s disease. Mol. Neurobiol. 2012, 46, 475–486. [Google Scholar] [CrossRef]
- Rodrigues, C.M.; Stieers, C.L.; Keene, C.D.; Ma, X.; Kren, B.T.; Low, W.C.; Steer, C.J. Tauroursodeoxycholic acid partially prevents apoptosis induced by 3-nitropropionic acid: Evidence for a mitochondrial pathway independent of the permeability transition. J. Neurochem. 2000, 75, 2368–2379. [Google Scholar] [CrossRef]
- Oveson, B.C.; Iwase, T.; Hackett, S.F.; Lee, S.Y.; Usui, S.; Sedlak, T.W.; Snyder, S.H.; Campochiaro, P.A.; Sung, J.U. Constituents of bile, bilirubin and TUDCA, protect against oxidative stress-induced retinal degeneration. J. Neurochem. 2011, 116, 144–153. [Google Scholar] [CrossRef] [Green Version]
- Drack, A.V.; Dumitrescu, A.V.; Bhattarai, S.; Gratie, D.; Stone, E.M.; Mullins, R.; Sheffield, V.C. TUDCA slows retinal degeneration in two different mouse models of retinitis pigmentosa and prevents obesity in Bardet-Biedl syndrome type 1 mice. Invest. Ophthalmol. Vis. Sci. 2012, 53, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Baehr, W.; Fu, Y. Chemical chaperone TUDCA preserves cone photoreceptors in a mouse model of Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2012, 53, 3349–3356. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.J.; Walker, T.A.; Choi, H.Y.; Faulkner, A.E.; Kim, M.K.; Sidney, S.S.; Boyd, A.P.; Nickerson, J.M.; Boatright, J.H.; Pardue, M.T. Tauroursodeoxycholic acid preservation of photoreceptor structure and function in the rd10 mouse through postnatal day 30. Invest. Ophthalmol. Vis. Sci. 2008, 49, 2148–2155. [Google Scholar] [CrossRef] [Green Version]
- Amaral, J.D.; Viana, R.J.; Ramalho, R.M.; Steer, C.J.; Rodrigues, C.M. Bile acids: Regulation of apoptosis by ursodeoxycholic acid. J. Lipid Res. 2009, 50, 1721–1734. [Google Scholar] [CrossRef] [Green Version]
- Uppala, J.K.; Gani, A.R.; Ramaiah, K.V.A. Chemical chaperone, TUDCA unlike PBA, mitigates protein aggregation efficiently and resists ER and non-ER stress induced HepG2 cell death. Sci. Rep. 2017, 7, 3831. [Google Scholar] [CrossRef]
- Xie, Q.; Khaoustov, V.I.; Chung, C.C.; Sohn, J.; Krishnan, B.; Lewis, D.E.; Yoffe, B. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology 2002, 36, 592–601. [Google Scholar] [CrossRef]
- Fonseca, I.; Gordino, G.; Moreira, S.; Nunes, M.J.; Azevedo, C.; Gama, M.J.; Rodrigues, E.; Rodrigues, C.M.P.; Castro-Caldas, M. Tauroursodeoxycholic Acid Protects Against Mitochondrial Dysfunction and Cell Death via Mitophagy in Human Neuroblastoma Cells. Mol. Neurobiol. 2017, 54, 6107–6119. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.M.; Sola, S.; Brito, M.A.; Brondino, C.D.; Brites, D.; Moura, J.J. Amyloid beta-peptide disrupts mitochondrial membrane lipid and protein structure: Protective role of tauroursodeoxycholate. Biochem. Biophys. Res. Commun. 2001, 281, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Huo, X.; Rezaei, D.; Zhang, Q.; Zhang, X.; Yu, C.; Asanuma, K.; Cheng, E.; Pham, T.H.; Wang, D.H.; et al. In Barrett’s esophagus patients and Barrett’s cell lines, ursodeoxycholic acid increases antioxidant expression and prevents DNA damage by bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, 129–139. [Google Scholar] [CrossRef]
- Moreira, S.; Fonseca, I.; Nunes, M.J.; Rosa, A.; Lemos, L.; Rodrigues, E.; Carvalho, A.N.; Outeiro, T.F.; Rodrigues, C.M.P.; Gama, M.J.; et al. Nrf2 activation by tauroursodeoxycholic acid in experimental models of Parkinson’s disease. Exp. Neurol. 2017, 295, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Alhasani, R.H.; Almarhoun, M.; Zhou, X.; Reilly, J.; Patterson, S.; Zeng, Z.; Shu, X. Tauroursodeoxycholic Acid Protects Retinal Pigment Epithelial Cells from Oxidative Injury and Endoplasmic Reticulum Stress In Vitro. Biomedicines 2020, 8, 367. [Google Scholar] [CrossRef]
- Ljubuncic, P.; Abu-Salach, O.; Bomzon, A. Ursodeoxycholic acid and superoxide anion. World J. Gastroenterol. 2005, 11, 4875–4878. [Google Scholar] [CrossRef]
- Lapenna, D.; Ciofani, G.; Festi, D.; Neri, M.; Pierdomenico, S.D.; Giamberardino, M.A.; Cuccurullo, F. Antioxidant properties of ursodeoxycholic acid. Biochem. Pharmacol. 2002, 64, 1661–1667. [Google Scholar] [CrossRef]
- Heuman, D.M.; Bajaj, R. Ursodeoxycholate conjugates protect against disruption of cholesterol-rich membranes by bile salts. Gastroenterology 1994, 106, 1333–1341. [Google Scholar] [CrossRef]
- Burns, M.E.; Arshavsky, V.Y. Beyond counting photons: Trials and trends in vertebrate visual transduction. Neuron 2005, 48, 387–401. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.-D.; Salinas, R.Y.; Arshavsky, V.Y. Discs of mammalian rod photoreceptors form through the membrane evagination mechanism. J. Cell Biol. 2015, 211, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Boesze-Battaglia, K.; Hennessey, T.; Albert, A.D. Cholesterol heterogeneity in bovine rod outer segment disk membranes. J. Biol. Chem. 1989, 264, 8151–8155. [Google Scholar] [CrossRef]
- Boesze-Battaglia, K.; Schimmel, R. Cell membrane lipid composition and distribution: Implications for cell function and lessons learned from photoreceptors and platelets. J. Exp. Biol. 1997, 200, 2927–2936. [Google Scholar] [CrossRef]
- Redmond, R.W.; Gamlin, J.N. A compilation of singlet oxygen yields from biologically relevant molecules. Photochem. Photobiol. 1999, 70, 391–475. [Google Scholar] [CrossRef]
- Bonnett, R.; McGarvey, D.J.; Harriman, A.; Land, E.J.; Truscott, T.G.; Winfield, U.J. Photophysical properties of meso-tetraphenylporphyrin and some meso-tetra(hydroxyphenyl)porphyrins. Photochem. Photobiol. 1988, 48, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Rozanowska, M.; Jarvis-Evans, J.; Korytowski, W.; Boulton, M.E.; Burke, J.M.; Sarna, T. Blue light-induced reactivity of retinal age pigment. In vitro generation of oxygen-reactive species. J. Biol. Chem. 1995, 270, 18825–18830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zadlo, A.; Burke, J.M.; Sarna, T. Effect of untreated and photobleached bovine RPE melanosomes on the photoinduced peroxidation of lipids. Photochem. Photobiol. Sci. 2009, 8, 830–837. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol an open–source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Seelig, J.; Waespe-Sarcevic, N. Molecular order in cis and trans unsaturated phospholipid bilayers. Biochemistry 1978, 17, 3310–3315. [Google Scholar] [CrossRef] [PubMed]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Markiewicz, M.; Pasenkiewicz-Gierula, M. Comparative Model Studies of Gastric Toxicity of Nonsteroidal Anti-Inflammatory Drugs. Langmuir 2011, 27, 6950–6961. [Google Scholar] [CrossRef] [PubMed]
- Pasenkiewicz-Gierula, M.; Róg, T.; Grochowski, J.; Serda, P.; Czarnecki, R.; Librowski, T.; Lochyński, S. Effects of a Carane Derivative Local Anesthetic on a Phospholipid Bilayer Studied by Molecular Dynamics Simulation. Biophys. J. 2003, 85, 1248–1258. [Google Scholar] [CrossRef] [Green Version]
- Gandhavadi, M.; Allende, D.; Vidal, A.; Simon, S.A.; McIntosh, T.J. Structure, composition, and peptide binding properties of detergent soluble bilayers and detergent resistant rafts. Biophys. J. 2002, 82, 1469–1482. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Ogilby, P.R. Singlet oxygen: There is indeed something new under the sun. Chem. Soc. Rev. 2010, 39, 3181–3209. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R. Photosensitized generation of singlet oxygen. Photochem. Photobiol. 2006, 82, 1161–1177. [Google Scholar] [CrossRef] [PubMed]
- Krasnovsky, A.A., Jr.; Kagan, V.E. Photosensitization and quenching of singlet oxygen by pigments and lipids of photoreceptor cells of the retina. FEBS Lett. 1979, 108, 152–154. [Google Scholar] [CrossRef] [Green Version]
- Rozanowska, M.; Sarna, T. Light-induced damage to the retina: Role of rhodopsin chromophore revisited. Photochem. Photobiol. 2005, 81, 1305–1330. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.Y.; Cringle, S.J.; Su, E.N. Intraretinal oxygen distribution in the monkey retina and the response to systemic hyperoxia. Invest. Ophthalmol. Vis. Sci. 2005, 46, 4728–4733. [Google Scholar] [CrossRef]
- Hao, W.; Wenzel, A.; Obin, M.S.; Chen, C.K.; Brill, E.; Krasnoperova, N.V.; Eversole-Cire, P.; Kleyner, Y.; Taylor, A.; Simon, M.I.; et al. Evidence for two apoptotic pathways in light-induced retinal degeneration. Nat. Genet. 2002, 32, 254–260. [Google Scholar] [CrossRef]
- Boulton, M.; Rózanowska, M.; Rózanowski, B. Retinal photodamage. J. Photochem. Photobiol. B 2001, 64, 144–161. [Google Scholar] [CrossRef]
- Maeda, T.; Golczak, M.; Maeda, A. Retinal photodamage mediated by all-trans-retinal. Photochem. Photobiol. 2012, 88, 1309–1319. [Google Scholar] [CrossRef] [Green Version]
- Saari, J.C.; Garwin, G.G.; van Hooser, J.P.; Palczewski, K. Reduction of all-trans-retinal limits regeneration of visual pigment in mice. Vision Res. 1998, 38, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Harper, W.S.; Gaillard, E.R. Studies of all-trans-retinal as a photooxidizing agent. Photochem. Photobiol. 2001, 73, 71–76. [Google Scholar] [CrossRef]
- Rozanowska, M.; Wessels, J.; Boulton, M.; Burke, J.M.; Rodgers, M.A.; Truscott, T.G.; Sarna, T. Blue light-induced singlet oxygen generation by retinal lipofuscin in non-polar media. Free Radic. Biol. Med. 1998, 24, 1107–1112. [Google Scholar] [CrossRef]
- Acar, N.; Berdeaux, O.; Gregoire, S.; Cabaret, S.; Martine, L.; Gain, P.; Thuret, G.; Creuzot-Garcher, C.P.; Bron, A.M.; Bretillon, L. Lipid composition of the human eye: Are red blood cells a good mirror of retinal and optic nerve fatty acids? PLoS ONE 2012, 7, e35102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koscielniak, A.; Serafin, M.; Duda, M.; Oles, T.; Zadlo, A.; Broniec, A.; Berdeaux, O.; Gregoire, S.; Bretillon, L.; Sarna, T.; et al. Oxidation-Induced Increase In Photoreactivity of Bovine Retinal Lipid Extract. Cell Biophys. 2017, 75, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Egorov, S.; Kurella, E.G.; Boldyrev, A.A.; Krasnovsky, A.A., Jr. Quenching of singlet molecular oxygen by carnosine and related antioxidants. Monitoring 1270-nm phosphorescence in aqueous media. Biochem. Mol. Biol. Int. 1997, 41, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, A.; Feitelson, J. Reactivity of singlet oxygen toward amino acids and peptides. Photochem. Photobiol. 1994, 59, 284–289. [Google Scholar] [CrossRef]
- Broniec, A.; Klosinski, R.; Pawlak, A.; Wrona-Krol, M.; Thompson, D.; Sarna, T. Interactions of plasmalogens and their diacyl analogs with singlet oxygen in selected model systems. Free Radic. Biol. Med. 2011, 50, 892–898. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.W.; Minotto, J.B.; de Oliveira, M.R.; Zanotto-Filho, A.; Behr, G.A.; Rocha, R.F.; Moreira, J.C.; Klamt, F. Scavenging and antioxidant potential of physiological taurine concentrations against different reactive oxygen/nitrogen species. Pharmacol. Rep. 2010, 62, 185–193. [Google Scholar] [CrossRef]
- Seidel, U.; Lüersen, K.; Huebbe, P.; Rimbach, G. Taurine Enhances Iron-Related Proteins and Reduces Lipid Peroxidation in Differentiated C2C12 Myotubes. Antioxidants 2020, 9, 1071. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, D.; Yi, B.; Liao, Z.; Tang, L.; Yin, D.; He, M. Taurine supplementation reduces oxidative stress and protects the liver in an iron-overload murine model. Mol. Med. Rep. 2014, 10, 2255–2262. [Google Scholar] [CrossRef] [Green Version]
- You, J.S.; Chang, K.J. Effects of taurine supplementation on lipid peroxidation, blood glucose and blood lipid metabolism in streptozotocin-induced diabetic rats. Adv. Exp. Med. Biol. 1998, 442, 163–168. [Google Scholar]
- DeLange, R.J.; Glazer, A.N. Bile acids: Antioxidants or enhancers of peroxidation depending on lipid concentration. Arch. Biochem. Biophys. 1990, 276, 19–25. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y. Tauroursodeoxycholic Acid Alleviates H(2)O(2)-Induced Oxidative Stress and Apoptosis via Suppressing Endoplasmic Reticulum Stress in Neonatal Rat Cardiomyocytes. Dose Response Publ. Int. Horm. Soc. 2018, 16, 1559325818782631. [Google Scholar] [CrossRef] [Green Version]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Mello-Vieira, J.; Sousa, T.; Coutinho, A.; Fedorov, A.; Lucas, S.D.; Moreira, R.; Castro, R.E.; Rodrigues, C.M.; Prieto, M.; Fernandes, F. Cytotoxic bile acids, but not cytoprotective species, inhibit the ordering effect of cholesterol in model membranes at physiologically active concentrations. Biochim. Biophys. Acta 2013, 1828, 2152–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, M.H.; Nash, Z.A.; Takemori, N.; Fliesler, S.J.; McClellan, M.E.; Naash, M.I. Differential distribution of proteins and lipids in detergent-resistant and detergent-soluble domains in rod outer segment plasma membranes and disks. J. Neurochem. 2008, 104, 336–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Blat, A.; Stepanenko, T.; Bulat, K.; Wajda, A.; Dybas, J.; Mohaissen, T.; Alcicek, F.C.; Szczesny-Malysiak, E.; Malek, K.; Fedorowicz, A.; et al. Spectroscopic Signature of Red Blood Cells in a D-Galactose-Induced Accelerated Aging Model. Int. J. Mol. Sci. 2021, 22, 2660. [Google Scholar] [CrossRef] [PubMed]
- Dybas, J.; Bulat, K.; Blat, A.; Mohaissen, T.; Wajda, A.; Mardyla, M.; Kaczmarska, M.; Franczyk-Zarow, M.; Malek, K.; Chlopicki, S.; et al. Age-related and atherosclerosis-related erythropathy in ApoE/LDLR(-/-) mice. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165972. [Google Scholar] [CrossRef] [PubMed]
- Blat, A.; Dybas, J.; Kaczmarska, M.; Chrabaszcz, K.; Bulat, K.; Kostogrys, R.B.; Cernescu, A.; Malek, K.; Marzec, K.M. An Analysis of Isolated and Intact RBC Membranes—A Comparison of a Semiquantitative Approach by Means of FTIR, Nano-FTIR, and Raman Spectroscopies. Anal. Chem. 2019, 91, 9867–9874. [Google Scholar] [CrossRef]
- Heraud, P.; Marzec, K.M.; Zhang, Q.H.; Yuen, W.S.; Carroll, J.; Wood, B.R. Label-free in vivo Raman microspectroscopic imaging of the macromolecular architecture of oocytes. Sci. Rep. 2017, 7, 8945. [Google Scholar] [CrossRef] [Green Version]
- Blat, A.; Dybas, J.; Chrabaszcz, K.; Bulat, K.; Jasztal, A.; Kaczmarska, M.; Pulyk, R.; Popiela, T.; Slowik, A.; Malek, K.; et al. FTIR, Raman and AFM characterization of the clinically valid biochemical parameters of the thrombi in acute ischemic stroke. Sci. Rep. 2019, 9, 15475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochan, K.; Chrabaszcz, K.; Szczur, B.; Maslak, E.; Dybas, J.; Marzec, K.M. IR and Raman imaging of murine brains from control and ApoE/LDLR−/− mice with advanced atherosclerosis. Analyst 2016, 141, 5329–5338. [Google Scholar] [CrossRef]
- Jamieson, L.E.; Wetherill, C.; Faulds, K.; Graham, D. Ratiometric Raman imaging reveals the new anti-cancer potential of lipid targeting drugs. Chem. Sci. 2018, 9, 6935–6943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirota, K.; Yagi, K.; Inaba, T.; Li, P.-C.; Murata, M.; Sugita, Y.; Kobayashi, T. Detection of Sphingomyelin Clusters by Raman Spectroscopy. Biophys. J. 2016, 111, 999–1007. [Google Scholar] [CrossRef] [Green Version]
- Marzec, K.M.; Wrobel, T.P.; Rygula, A.; Maslak, E.; Jasztal, A.; Fedorowicz, A.; Chlopicki, S.; Baranska, M. Visualization of the biochemical markers of atherosclerotic plaque with the use of Raman, IR and AFM. J. Biophotonics 2014, 7, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Maiti, N.; Thomas, S.; Debnath, A.; Kapoor, S. Raman and XPS study on the interaction of taurine with silver nanoparticles. RSC Adv. 2016, 6, 56406–56411. [Google Scholar] [CrossRef]
- Lima, R.J.C.; Freire, P.T.C.; Sasaki, J.M.; Melo, F.E.A.; Mendes Filho, J.; Moreira, R.L. Temperature-dependent Raman study of taurine single crystal. J. Raman Spectrosc. 2001, 32, 751–756. [Google Scholar] [CrossRef]
- Wisniewska, A.; Subczynski, W.K. Distribution of macular xanthophylls between domains in a model of photoreceptor outer segment membranes. Free Radic. Biol. Med. 2006, 41, 1257–1265. [Google Scholar] [CrossRef]
- Wisniewska, A.; Subczynski, W.K. Accumulation of macular xanthophylls in unsaturated membrane domains. Free Radic. Biol. Med. 2006, 40, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska-Becker, A.; Nawrocki, G.; Duda, M.; Subczynski, W.K. Structural aspects of the antioxidant activity of lutein in a model of photoreceptor membranes. Acta Biochim. Pol. 2012, 59, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subczynski, W.; Wisniewska-Becker, A.; Widomska, J. Xanthophyll–membrane interactions. In Carotenoids and Retinal Disease; Landrum, J.T., Nolan, J.M., Eds.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Polozova, A.; Litman, B.J. Cholesterol dependent recruitment of di22:6-PC by a G protein-coupled receptor into lateral domains. Biophys. J. 2000, 79, 2632–2643. [Google Scholar] [CrossRef] [Green Version]
- Lobysheva, E.; Taylor, C.M.; Marshall, G.R.; Kisselev, O.G. Tauroursodeoxycholic acid binds to the G-protein site on light activated rhodopsin. Exp. Eye Res. 2018, 170, 51–57. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Wisniewska, A.; Yin, J.-J.; Hyde, J.S.; Kusumi, A. Hydrophobic Barriers of Lipid Bilayer Membranes Formed by Reduction of Water Penetration by Alkyl Chain Unsaturation and Cholesterol. Biochemistry 1994, 33, 7670–7681. [Google Scholar] [CrossRef] [PubMed]
- Griffith, O.H.; Dehlinger, P.J.; Van, S.P. Shape of the hydrophobic barrier of phospholipid bilayers (Evidence for water penetration in biological membranes). J. Membr. Biol. 1974, 15, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska, A.; Subczynski, W.K. Effects of polar carotenoids on the shape of the hydrophobic barrier of phospholipid bilayers. Biochim. Biophys. Acta Biomembr. 1998, 1368, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Wisniewska, A.; Subczynski, W.K. Spin-label study on gramicidin–phosphatidylcholine interface: Fluidity, hydrophobicity and ion penetration. Curr. Top. Biophys. 1996, 20, 86–92. [Google Scholar]
- Duda, M.; Kawula, K.; Pawlak, A.; Sarna, T.; Wisniewska-Becker, A. EPR Studies on the Properties of Model Photoreceptor Membranes Made of Natural and Synthetic Lipids. Cell Biochem. Biophys. 2017, 75, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Duda, M.; Cygan, K.; Wisniewska-Becker, A. Effects of Curcumin on Lipid Membranes: An EPR Spin-label Study. Cell Biochem. Biophys. 2020, 78, 139–147. [Google Scholar] [CrossRef]
- Marsh, D. Electron spin resonance: Spin labels. In Membrane Spectroscopy; Grell, E., Ed.; Springer: Berlin/Heidelberg, Germany, 1981; pp. 51–142. [Google Scholar]
- Pasenkiewicz-Gierula, M.; Subczynski, W.K.; Kusumi, A. Rotational diffusion of a steroid molecule in phosphatidylcholine-cholesterol membranes: Fluid-phase microimmiscibility in unsaturated phosphatidylcholine-cholesterol membranes. Biochemistry 1990, 29, 4059–4069. [Google Scholar] [CrossRef]
- Kusumi, A.; Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Hyde, J.S.; Merkle, H. Spin-label studies on phosphatidylcholine-cholesterol membranes: Effects of alkyl chain length and unsaturation in the fluid phase. Biochim. Biophys. Acta 1986, 854, 307–317. [Google Scholar] [CrossRef]
- Arai, Y.; Choi, B.; Kim, B.J.; Rim, W.; Park, S.; Park, H.; Ahn, J.; Lee, S.-H. Tauroursodeoxycholic acid (TUDCA) counters osteoarthritis by regulating intracellular cholesterol levels and membrane fluidity of degenerated chondrocytes. Biomater. Sci. 2019, 7, 3178–3189. [Google Scholar] [CrossRef] [PubMed]








| Compound | Rate constants for Interactions of 1O2 (M−1s−1) | Solvent |
|---|---|---|
| TUDCA | (1.98 ± 0.32) × 105 | DMSO-d6:CHCl3 mixture (1:1, v/v) |
| UDCA | (0.39 ± 0.13) × 105 | DMSO-d6:CHCl3 mixture (1:1, v/v) |
| Taurine a | <3.00 × 103 | Deuterated water (D2O) |
| Phospholipids b | 0.8 ÷ 1.8 × 105 | Carbon tetrachloride (CCl4) |
| Cholesterol c | 5.7 ×·104 | Benzene |
| a–Tocopherol d | (1.21 ± 0.17) × 108 | Carbon tetrachloride (CCl4) |
| TUDCA Molecule | |||||
|---|---|---|---|---|---|
| Sulphate Group OS | Amide Group NH | Carbonyl Group OE1 | Hydroxyl Group at the 3rd Carbon Atom of the Sterol Ring OH3 | Hydroxyl Group at the 7th Carbon Atom of the Sterol Ring OH7 | |
| POPC | — | 0.03 ± 0.05 | — | 0.4 ± 0.14 | 0.35 ± 0.14 |
| Phosphate group OP | — | 0.01 ± 0.03 | — | 0.07 ± 0.08 | 0.15 ± 0.1 |
| Carbonyl group OC | — | 0.03 ± 0.05 | — | 0.33 ± 0.14 | 0.19 ± 0.13 |
| WATER | 4.93 ± 0.42 | 0.19 ± 0.12 | 0.87 ± 0.19 | 1.02 ± 0.28 | 0.98 ± 0.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabat, M.J.; Wiśniewska-Becker, A.M.; Markiewicz, M.; Marzec, K.M.; Dybas, J.; Furso, J.; Pabisz, P.; Duda, M.; Pawlak, A.M. Tauroursodeoxycholic Acid (TUDCA)—Lipid Interactions and Antioxidant Properties of TUDCA Studied in Model of Photoreceptor Membranes. Membranes 2021, 11, 327. https://0-doi-org.brum.beds.ac.uk/10.3390/membranes11050327
Sabat MJ, Wiśniewska-Becker AM, Markiewicz M, Marzec KM, Dybas J, Furso J, Pabisz P, Duda M, Pawlak AM. Tauroursodeoxycholic Acid (TUDCA)—Lipid Interactions and Antioxidant Properties of TUDCA Studied in Model of Photoreceptor Membranes. Membranes. 2021; 11(5):327. https://0-doi-org.brum.beds.ac.uk/10.3390/membranes11050327
Chicago/Turabian StyleSabat, Michał J., Anna M. Wiśniewska-Becker, Michał Markiewicz, Katarzyna M. Marzec, Jakub Dybas, Justyna Furso, Paweł Pabisz, Mariusz Duda, and Anna M. Pawlak. 2021. "Tauroursodeoxycholic Acid (TUDCA)—Lipid Interactions and Antioxidant Properties of TUDCA Studied in Model of Photoreceptor Membranes" Membranes 11, no. 5: 327. https://0-doi-org.brum.beds.ac.uk/10.3390/membranes11050327