Modelling Nuclear Morphology and Shape Transformation: A Review
Abstract
:1. Introduction
2. Biophysical Elements Involved in Regulating Nuclear Morphology
2.1. Internal Structure of the Nucleus
2.2. Cytoskeleton
2.3. Extracellular Matrix and Cell Adhesion
2.4. Physical Confinement
2.5. Osmolarity
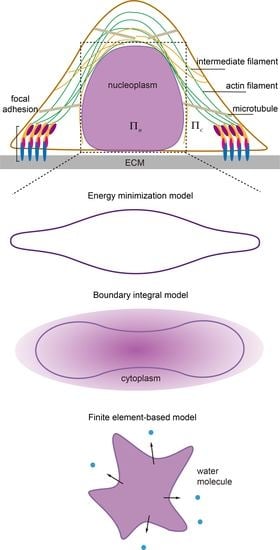
3. Continuum Models for Describing Nuclear Morphology
3.1. Energy Minimization Model
3.2. Boundary Integral Model
3.3. Finite Element-Based Models
4. Conclusions and Outlooks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, L.; Schwartz, C.; Magidson, V.; Khodjakov, A.; Oliferenko, S. The spindle pole bodies facilitate nuclear envelope division during closed mitosis in fission yeast. PLoS Biol. 2007, 5, e170. [Google Scholar] [CrossRef]
- Gonzalez, Y.; Meerbrey, K.; Chong, J.; Torii, Y.; Padte, N.N.; Sazer, S. Nuclear shape, growth and integrity in the closed mitosis of fission yeast depend on the Ran-GTPase system, the spindle pole body and the endoplasmic reticulum. J. Cell Sci. 2009, 122, 2464–2472. [Google Scholar] [CrossRef] [Green Version]
- Yam, C.; He, Y.; Zhang, D.; Chiam, K.-H.; Oliferenko, S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr. Biol. 2011, 21, 1314–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-K.; Louhghalam, A.; Lee, G.; Schafer, B.W.; Wirtz, D.; Kim, D.-H. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Vishavkarma, R.; Raghavan, S.; Kuyyamudi, C.; Majumder, A.; Dhawan, J.; Pullarkat, P.A. Role of actin filaments in correlating nuclear shape and cell spreading. PLoS ONE 2014, 9, e107895. [Google Scholar]
- Lovett, D.B.; Shekhar, N.; Nickerson, J.A.; Roux, K.J.; Lele, T.P. Modulation of nuclear shape by substrate rigidity. Cell. Mol. Bioeng. 2013, 6, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Khatau, S.B.; Hale, C.M.; Stewart-Hutchinson, P.J.; Patel, M.S.; Stewart, C.L.; Searson, P.C.; Hodzic, D.; Wirtz, D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA 2009, 106, 19017–19022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, P.M.; Denais, C.; Bakshi, M.C.; Lammerding, J. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 2014, 7, 293–306. [Google Scholar] [CrossRef] [Green Version]
- Giverso, C.; Grillo, A.; Preziosi, L. Influence of nucleus deformability on cell entry into cylindrical structures. Biomech. Model. Mechanobiol. 2014, 13, 481–502. [Google Scholar] [CrossRef] [Green Version]
- Lomakin, A.; Cattin, C.; Cuvelier, D.; Alraies, Z.; Molina, M.; Nader, G.; Srivastava, N.; Saez, P.; Garcia-Arcos, J.; Zhitnyak, I. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 2020, 370, eaba2894. [Google Scholar] [CrossRef]
- Venturini, V.; Pezzano, F.; Castro, F.C.; Häkkinen, H.-M.; Jiménez-Delgado, S.; Colomer-Rosell, M.; Marro, M.; Tolosa-Ramon, Q.; Paz-López, S.; Valverde, M.A. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science 2020, 370, eaba2644. [Google Scholar] [CrossRef]
- Lee, H.-P.; Alisafaei, F.; Adebawale, K.; Chang, J.; Shenoy, V.B.; Chaudhuri, O. The nuclear piston activates mechanosensitive ion channels to generate cell migration paths in confining microenvironments. Sci. Adv. 2021, 7, eabd4058. [Google Scholar] [CrossRef]
- Sazer, S.; Lynch, M.; Needleman, D. Deciphering the evolutionary history of open and closed mitosis. Curr. Biol. 2014, 24, R1099–R1103. [Google Scholar] [CrossRef] [Green Version]
- Smoyer, C.J.; Jaspersen, S.L. Breaking down the wall: The nuclear envelope during mitosis. Curr. Opin. Cell Biol. 2014, 26, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Dey, G.; Culley, S.; Curran, S.; Schmidt, U.; Henriques, R.; Kukulski, W.; Baum, B. Closed mitosis requires local disassembly of the nuclear envelope. Nature 2020, 585, 119–123. [Google Scholar] [CrossRef]
- Qu, L.-H.; Sun, M.-X. The plant cell nucleus is constantly alert and highly sensitive to repetitive local mechanical stimulations. Plant Cell Rep. 2007, 26, 1187–1193. [Google Scholar] [CrossRef]
- Xiong, T.C.; Jauneau, A.; Ranjeva, R.; Mazars, C. Isolated plant nuclei as mechanical and thermal sensors involved in calcium signalling. Plant J. 2004, 40, 12–21. [Google Scholar] [CrossRef]
- Kirby, T.J.; Lammerding, J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 2018, 20, 373–381. [Google Scholar] [CrossRef]
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017, 216, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Enyedi, B.; Jelcic, M.; Niethammer, P. The cell nucleus serves as a mechanotransducer of tissue damage-induced inflammation. Cell 2016, 165, 1160–1170. [Google Scholar] [CrossRef] [Green Version]
- Enyedi, B.; Niethammer, P. Nuclear membrane stretch and its role in mechanotransduction. Nucleus 2017, 8, 156–161. [Google Scholar] [CrossRef] [Green Version]
- Uhler, C.; Shivashankar, G. Nuclear mechanopathology and cancer diagnosis. Trends Cancer 2018, 4, 320–331. [Google Scholar] [CrossRef] [Green Version]
- Maurer, M.; Lammerding, J. The driving force: Nuclear mechanotransduction in cellular function, fate, and disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Uhler, C.; Shivashankar, G. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017, 18, 717–727. [Google Scholar] [CrossRef]
- Jain, N.; Iyer, K.V.; Kumar, A.; Shivashankar, G. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. USA 2013, 110, 11349–11354. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Marcel, N.; Sarin, A.; Shivashankar, G. Role of actin dependent nuclear deformation in regulating early gene expression. PLoS ONE 2012, 7, e53031. [Google Scholar] [CrossRef] [Green Version]
- Obara, K.; Kuriyama, H.; Fukuda, H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 2001, 125, 615–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kost, B.; Chua, N.-H. The plant cytoskeleton: Vacuoles and cell walls make the difference. Cell 2002, 108, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Zwerger, M.; Ho, C.Y.; Lammerding, J. Nuclear mechanics in disease. Annu. Rev. Biomed. Eng. 2011, 13, 397–428. [Google Scholar] [CrossRef] [Green Version]
- De Sandre-Giovannoli, A.; Bernard, R.; Cau, P.; Navarro, C.; Amiel, J.; Boccaccio, I.; Lyonnet, S.; Stewart, C.L.; Munnich, A.; Le Merrer, M. Lamin a truncation in Hutchinson-Gilford progeria. Science 2003, 300, 2055. [Google Scholar] [CrossRef]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef] [Green Version]
- Goldman, R.D.; Shumaker, D.K.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Gordon, L.B.; Gruenbaum, Y.; Khuon, S.; Mendez, M.; Varga, R. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8963–8968. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Zhou, Z. Genetics of aging, progeria and lamin disorders. Curr. Opin. Genet. Dev. 2014, 26, 41–46. [Google Scholar] [CrossRef]
- Lele, T.P.; Dickinson, R.B.; Gundersen, G.G. Mechanical principles of nuclear shaping and positioning. J. Cell Biol. 2018, 217, 3330–3342. [Google Scholar] [CrossRef] [Green Version]
- Dahl, K.N.; Scaffidi, P.; Islam, M.F.; Yodh, A.G.; Wilson, K.L.; Misteli, T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson–Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2006, 103, 10271–10276. [Google Scholar] [CrossRef] [Green Version]
- Bione, S.; Maestrini, E.; Rivella, S.; Mancini, M.; Regis, S.; Romeo, G.; Toniolo, D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994, 8, 323–327. [Google Scholar] [CrossRef]
- Rowat, A.; Lammerding, J.; Ipsen, J.H. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys. J. 2006, 91, 4649–4664. [Google Scholar] [CrossRef] [Green Version]
- Lammerding, J.; Hsiao, J.; Schulze, P.C.; Kozlov, S.; Stewart, C.L.; Lee, R.T. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005, 170, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Tseng, Y.; Lee, J.S.; Kole, T.P.; Jiang, I.; Wirtz, D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J. Cell Sci. 2004, 117, 2159–2167. [Google Scholar] [CrossRef] [Green Version]
- Lherbette, M.; Dos Santos, Á.; Hari-Gupta, Y.; Fili, N.; Toseland, C.P.; Schaap, I.A. Atomic Force Microscopy micro-rheology reveals large structural inhomogeneities in single cell-nuclei. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guilak, F.; Tedrow, J.R.; Burgkart, R. Viscoelastic properties of the cell nucleus. Biochem. Biophys. Res. Commun. 2000, 269, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, S.; Maeda, K.; Ohashi, T.; Sato, M. Flow-induced hardening of endothelial nucleus as an intracellular stress-bearing organelle. J. Biomech. 2005, 38, 1751–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowat, A.C.; Foster, L.J.; Nielsen, M.M.; Weiss, M.; Ipsen, J.H. Characterization of the elastic properties of the nuclear envelope. J. R. Soc. Interface 2005, 2, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Pajerowski, J.D.; Dahl, K.N.; Zhong, F.L.; Sammak, P.J.; Discher, D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 15619–15624. [Google Scholar] [CrossRef]
- Caille, N.; Thoumine, O.; Tardy, Y.; Meister, J.-J. Contribution of the nucleus to the mechanical properties of endothelial cells. J. Biomech. 2002, 35, 177–187. [Google Scholar] [CrossRef]
- Stephens, A.D.; Banigan, E.J.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 2017, 28, 1984–1996. [Google Scholar] [CrossRef]
- Wei, F.; Lan, F.; Liu, B.; Liu, L.; Li, G. Poroelasticity of cell nuclei revealed through atomic force microscopy characterization. Appl. Phys. Lett. 2016, 109, 213701. [Google Scholar] [CrossRef]
- Hobson, C.M.; Kern, M.; O’Brien, E.T.; Stephens, A.D.; Falvo, M.R.; Superfine, R. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 2020, 31, 1788–1801. [Google Scholar] [CrossRef]
- Liang, L.; Wang, X.; Da, X.; Chen, T.; Chen, W.R. Noninvasive determination of cell nucleoplasmic viscosity by fluorescence correlation spectroscopy. J. Biomed. Opt. 2009, 14, 024013. [Google Scholar] [CrossRef]
- Zhang, J.; Alisafaei, F.; Nikolić, M.; Nou, X.A.; Kim, H.; Shenoy, V.B.; Scarcelli, G. Nuclear mechanics within intact cells is regulated by cytoskeletal network and internal nanostructures. Small 2020, 16, 1907688. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.M.; Koo, P.K.; Zhao, Y.; Mochrie, S.G.; King, M.C. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 2015, 6, 7159. [Google Scholar] [CrossRef] [Green Version]
- Khodjakov, A.; La Terra, S.; Chang, F. Laser microsurgery in fission yeast: Role of the mitotic spindle midzone in anaphase B. Curr. Biol. 2004, 14, 1330–1340. [Google Scholar] [CrossRef] [Green Version]
- Vaziri, A.; Mofrad, M.R.K. Mechanics and deformation of the nucleus in micropipette aspiration experiment. J. Biomech. 2007, 40, 2053–2062. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, A.; Lee, H.; Mofrad, M.K. Deformation of the cell nucleus under indentation: Mechanics and mechanisms. J. Mater. Res. 2006, 21, 2126–2135. [Google Scholar] [CrossRef]
- Li, Y.; Lovett, D.; Zhang, Q.; Neelam, S.; Kuchibhotla, R.A.; Zhu, R.; Gundersen, G.G.; Lele, T.P.; Dickinson, R.B. Moving cell boundaries drive nuclear shaping during cell spreading. Biophys. J. 2015, 109, 670–686. [Google Scholar] [CrossRef] [Green Version]
- Versaevel, M.; Grevesse, T.; Gabriele, S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 2012, 3, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hobson, C.M.; Stephens, A.D. Modeling of Cell Nuclear Mechanics: Classes, Components, and Applications. Cells 2020, 9, 1623. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science: New York, NY, USA, 2015; p. 180. [Google Scholar]
- Franke, W.W.; Scheer, U.; Krohne, G.; Jarasch, E.-D. The nuclear envelope and the architecture of the nuclear periphery. J. Cell Biol. 1981, 91, 39s–50s. [Google Scholar] [CrossRef]
- D’angelo, M.A.; Anderson, D.J.; Richard, E.; Hetzer, M.W. Nuclear pores form de novo from both sides of the nuclear envelope. Science 2006, 312, 440–443. [Google Scholar] [CrossRef] [Green Version]
- De Magistris, P.; Antonin, W. The dynamic nature of the nuclear envelope. Curr. Biol. 2018, 28, R487–R497. [Google Scholar] [CrossRef] [Green Version]
- Senda, T.; Iizuka-Kogo, A.; Shimomura, A. Visualization of the nuclear lamina in mouse anterior pituitary cells and immunocytochemical detection of lamin A/C by quick-freeze freeze-substitution electron microscopy. J. Histochem. Cytochem. 2005, 53, 497–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mans, B.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 2004, 3, 1625–1650. [Google Scholar] [CrossRef] [Green Version]
- Jahed, Z.; Mofrad, M.R. The nucleus feels the force, LINCed in or not! Curr. Opin. Cell Biol. 2019, 58, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Lele, T.P. Mechanics of nuclear membranes. J. Cell Sci. 2019, 132, jcs229245. [Google Scholar] [CrossRef] [Green Version]
- Jevtić, P.; Edens, L.J.; Vuković, L.D.; Levy, D.L. Sizing and shaping the nucleus: Mechanisms and significance. Curr. Opin. Cell Biol. 2014, 28, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Chalut, K.J.; Höpfler, M.; Lautenschläger, F.; Boyde, L.; Chan, C.J.; Ekpenyong, A.; Martinez-Arias, A.; Guck, J. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys. J. 2012, 103, 2060–2070. [Google Scholar] [CrossRef] [Green Version]
- Furusawa, T.; Rochman, M.; Taher, L.; Dimitriadis, E.K.; Nagashima, K.; Anderson, S.; Bustin, M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 2015, 6, 6138. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Ragnauth, C.; Greener, M.J.; Shanahan, C.M.; Roberts, R.G. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics 2002, 80, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Dahl, K.N.; Ribeiro, A.J.; Lammerding, J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008, 102, 1307–1318. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Beck, K.A. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J. Cell Sci. 2004, 117, 619–629. [Google Scholar] [CrossRef] [Green Version]
- Wilhelmsen, K.; Litjens, S.H.; Kuikman, I.; Tshimbalanga, N.; Janssen, H.; van den Bout, I.; Raymond, K.; Sonnenberg, A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 2005, 171, 799–810. [Google Scholar] [CrossRef]
- Sun, D.; Leung, C.L.; Liem, R. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): Identification of a novel group of microtubule associated proteins. J. Cell Sci. 2001, 114, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Wiche, G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998, 111, 2477–2486. [Google Scholar] [CrossRef]
- Zhao, T.; Graham, O.S.; Raposo, A.; St Johnston, D. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 2012, 336, 999–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampoelz, B.; Azou-Gros, Y.; Fabre, R.; Markova, O.; Puech, P.-H.; Lecuit, T. Microtubule-induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development 2011, 138, 3377–3386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerlitz, G.; Reiner, O.; Bustin, M. Microtubule dynamics alter the interphase nucleus. Cell. Mol. Life Sci. 2013, 70, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Li, B.; Si, F.; Phillip, J.M.; Wirtz, D.; Sun, S.X. Volume regulation and shape bifurcation in the cell nucleus. J. Cell Sci. 2015, 128, 3375–3385. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Cho, S.; Wirtz, D. Tight coupling between nucleus and cell migration through the perinuclear actin cap. J. Cell Sci. 2014, 127, 2528–2541. [Google Scholar]
- Kim, D.-H.; Chambliss, A.B.; Wirtz, D. The multi-faceted role of the actin cap in cellular mechanosensation and mechanotransduction. Soft Matter 2013, 9, 5516–5523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patteson, A.E.; Vahabikashi, A.; Pogoda, K.; Adam, S.A.; Mandal, K.; Kittisopikul, M.; Sivagurunathan, S.; Goldman, A.; Goldman, R.D.; Janmey, P.A. Vimentin protects cells against nuclear rupture and DNA damage during migration. J. Cell Biol. 2019, 218, 4079–4092. [Google Scholar] [CrossRef] [PubMed]
- Lowery, J.; Kuczmarski, E.R.; Herrmann, H.; Goldman, R.D. Intermediate filaments play a pivotal role in regulating cell architecture and function. J. Biol. Chem. 2015, 290, 17145–17153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarria, A.J.; Lieber, J.G.; Nordeen, S.K.; Evans, R.M. The presence or absence of a vimentin-type intermediate filament network affects the shape of the nucleus in human SW-13 cells. J. Cell Sci. 1994, 107, 1593–1607. [Google Scholar] [CrossRef]
- Trichet, L.; Le Digabel, J.; Hawkins, R.J.; Vedula, S.R.K.; Gupta, M.; Ribrault, C.; Hersen, P.; Voituriez, R.; Ladoux, B. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc. Natl. Acad. Sci. USA 2012, 109, 6933–6938. [Google Scholar] [CrossRef] [Green Version]
- Elosegui-Artola, A.; Oria, R.; Chen, Y.; Kosmalska, A.; Pérez-González, C.; Castro, N.; Zhu, C.; Trepat, X.; Roca-Cusachs, P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 2016, 18, 540. [Google Scholar] [CrossRef] [Green Version]
- Lo, C.-M.; Wang, H.-B.; Dembo, M.; Wang, Y.-l. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000, 79, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.; Bacakova, L.; Newman, C.; Hategan, A.; Griffin, M.; Discher, D. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 2004, 86, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Prager-Khoutorsky, M.; Lichtenstein, A.; Krishnan, R.; Rajendran, K.; Mayo, A.; Kam, Z.; Geiger, B.; Bershadsky, A.D. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 2011, 13, 1457–1465. [Google Scholar] [CrossRef]
- Doss, B.L.; Pan, M.; Gupta, M.; Grenci, G.; Mège, R.-M.; Lim, C.T.; Sheetz, M.P.; Voituriez, R.; Ladoux, B. Cell response to substrate rigidity is regulated by active and passive cytoskeletal stress. Proc. Natl. Acad. Sci. USA 2020, 117, 12817–12825. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Janmey, P.; Wang, Y.-l. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, M.; Racine, V.; Piel, M.; Pépin, A.; Dimitrov, A.; Chen, Y.; Sibarita, J.-B.; Bornens, M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 2006, 103, 19771–19776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanchanawong, P.; Shtengel, G.; Pasapera, A.M.; Ramko, E.B.; Davidson, M.W.; Hess, H.F.; Waterman, C.M. Nanoscale architecture of integrin-based cell adhesions. Nature 2010, 468, 580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupin, I.; Camand, E.; Etienne-Manneville, S. Classical cadherins control nucleus and centrosome position and cell polarity. J. Cell Biol. 2009, 185, 779–786. [Google Scholar] [CrossRef] [Green Version]
- Arias-Garcia, M.; Rickman, R.; Sero, J.; Yuan, Y.; Bakal, C. The cell-cell adhesion protein JAM3 determines nuclear deformability by regulating microtubule organization. bioRxiv 2020, 689737. [Google Scholar] [CrossRef] [Green Version]
- Wolf, K.; Te Lindert, M.; Krause, M.; Alexander, S.; Te Riet, J.; Willis, A.L.; Hoffman, R.M.; Figdor, C.G.; Weiss, S.J.; Friedl, P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013, 201, 1069–1084. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Moeendarbary, E.; Isermann, P.; Davidson, P.M.; Wang, X.; Chen, M.B.; Burkart, A.K.; Lammerding, J.; Kamm, R.D.; Shenoy, V.B. A chemomechanical model for nuclear morphology and stresses during cell transendothelial migration. Biophys. J. 2016, 111, 1541–1552. [Google Scholar] [CrossRef] [Green Version]
- Heo, S.-J.; Song, K.H.; Thakur, S.; Miller, L.M.; Cao, X.; Peredo, A.P.; Seiber, B.N.; Qu, F.; Driscoll, T.P.; Shenoy, V.B. Nuclear softening expedites interstitial cell migration in fibrous networks and dense connective tissues. Sci. Adv. 2020, 6, eaax5083. [Google Scholar] [CrossRef]
- Pullarkat, P.A.; Dommersnes, P.; Fernández, P.; Joanny, J.-F.; Ott, A. Osmotically driven shape transformations in axons. Phys. Rev. Lett. 2006, 96, 048104. [Google Scholar] [CrossRef] [Green Version]
- Hui, T.H.; Kwan, K.W.; Yip, T.T.C.; Fong, H.W.; Ngan, K.C.; Yu, M.; Yao, S.; Ngan, A.H.W.; Lin, Y. Regulating the Membrane Transport Activity and Death of Cells via Electroosmotic Manipulation. Biophys. J. 2016, 110, 2769–2778. [Google Scholar] [CrossRef] [Green Version]
- Stroka, K.M.; Jiang, H.; Chen, S.-H.; Tong, Z.; Wirtz, D.; Sun, S.X.; Konstantopoulos, K. Water permeation drives tumor cell migration in confined microenvironments. Cell 2014, 157, 611–623. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Jiang, H. Shape and dynamics of adhesive cells: Mechanical response of open systems. Phys. Rev. Lett. 2017, 118, 208102. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, L.; Mori, Y.; Sun, S.X. On the energy efficiency of cell migration in diverse physical environments. Proc. Natl. Acad. Sci. USA 2019, 116, 23894–23900. [Google Scholar] [CrossRef] [Green Version]
- Finan, J.D.; Chalut, K.J.; Wax, A.; Guilak, F. Nonlinear osmotic properties of the cell nucleus. Ann. Biomed. Eng. 2009, 37, 477. [Google Scholar] [CrossRef] [PubMed]
- Canham, P.B. The minimum energy of bending as a possible explanation of the biconcave shape of the human red blood cell. J. Theor. Biol. 1970, 26, 61–81. [Google Scholar] [CrossRef]
- Helfrich, W. Elastic properties of lipid bilayers: Theory and possible experiments. Z. Naturforsch. C 1973, 28, 693–703. [Google Scholar] [CrossRef]
- Zhu, Q.; Zheng, F.; Liu, A.P.; Qian, J.; Fu, C.; Lin, Y. Shape Transformation of the Nuclear Envelope during Closed Mitosis. Biophys. J. 2016, 111, 2309–2316. [Google Scholar] [CrossRef] [Green Version]
- Castagnetti, S.; Božič, B.; Svetina, S. Mechanical and molecular basis for the symmetrical division of the fission yeast nuclear envelope. Phys. Chem. Chem. Phys. 2015, 17, 15629–15636. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.W.G.; Huber, G.; Torii, Y.; Hirata, A.; Miller, J.; Sazer, S. Vesicle-like biomechanics governs important aspects of nuclear geometry in fission yeast. PLoS ONE 2007, 2, e948. [Google Scholar] [CrossRef]
- Emsellem, V.; Cardoso, O.; Tabeling, P. Vesicle deformation by microtubules: A phase diagram. Phys. Rev. E 1998, 58, 4807. [Google Scholar] [CrossRef]
- Seifert, U. Configurations of fluid membranes and vesicles. Adv. Phys. 1997, 46, 13–137. [Google Scholar] [CrossRef]
- Evans, E.A. Minimum energy analysis of membrane deformation applied to pipet aspiration and surface adhesion of red blood cells. Biophys. J. 1980, 30, 265–284. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, Y.; Saito, A.; Sazer, S. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus 2012, 3, 60–76. [Google Scholar] [CrossRef] [Green Version]
- King, M.C.; Lusk, C.; Blobel, G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 2006, 442, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Torbati, M.; Lele, T.P.; Agrawal, A. Ultradonut topology of the nuclear envelope. Proc. Natl. Acad. Sci. USA 2016, 113, 11094–11099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noguchi, H. Construction of nuclear envelope shape by a high-genus vesicle with pore-size constraint. Biophys. J. 2016, 111, 824–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, C.; Zheng, F.; Yao, J.; Wei, X.; Fu, C.; Shi, X.; Lin, Y. A Model for Bridging Microtubule Dynamics with Nuclear Envelope Shape Evolution during Closed Mitosis. J. Mech. Phys. Solids 2020, 144, 104116. [Google Scholar] [CrossRef]
- Daniels, B.R.; Masi, B.C.; Wirtz, D. Probing single-cell micromechanics in vivo: The microrheology of C. elegans developing embryos. Biophys. J. 2006, 90, 4712–4719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirtz, D. Particle-tracking microrheology of living cells: Principles and applications. Annu. Rev. Biophys. 2009, 38, 301–326. [Google Scholar] [CrossRef] [Green Version]
- Doubrovinski, K.; Swan, M.; Polyakov, O.; Wieschaus, E.F. Measurement of cortical elasticity in Drosophila melanogaster embryos using ferrofluids. Proc. Natl. Acad. Sci. USA 2017, 114, 1051–1056. [Google Scholar] [CrossRef] [Green Version]
- de Vries, A.H.; Krenn, B.E.; van Driel, R.; Subramaniam, V.; Kanger, J.S. Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett. 2007, 7, 1424–1427. [Google Scholar] [CrossRef]
- Celedon, A.; Hale, C.M.; Wirtz, D. Magnetic manipulation of nanorods in the nucleus of living cells. Biophys. J. 2011, 101, 1880–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caragine, C.M.; Haley, S.C.; Zidovska, A. Surface fluctuations and coalescence of nucleolar droplets in the human cell nucleus. Phys. Rev. Lett. 2018, 121, 148101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Power, H.; Wrobel, L.C. Boundary Integral Methods in Fluid Mechanics; Computational Mechanics: Southampton, UK, 1995; pp. 247–250. [Google Scholar]
- Pozrikidis, C. Boundary Integral and Singularity Methods for Linearized Viscous Flow; Cambridge University Press: New York, NY, USA, 1992; pp. 143–144. [Google Scholar]
- Brust-Mascher, I.; Civelekoglu-Scholey, G.; Kwon, M.; Mogilner, A.; Scholey, J.M. Model for anaphase B: Role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA 2004, 101, 15938–15943. [Google Scholar] [CrossRef] [Green Version]
- Brust-Mascher, I.; Scholey, J.M. Mitotic motors and chromosome segregation: The mechanism of anaphase B. Biochem. Soc. Trans. 2011, 39, 1149–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pozrikidis, C. Effect of membrane bending stiffness on the deformation of capsules in simple shear flow. J. Fluid Mech. 2001, 440, 269–291. [Google Scholar] [CrossRef]
- Hill, A.V. The heat of shortening and the dynamic constants of muscle. Proc. Royal Soc. B 1938, 126, 136–195. [Google Scholar]
- Deveraux, S.; Allena, R.; Aubry, D. A numerical model suggests the interplay between nuclear plasticity and stiffness during a perfusion assay. J. Theor. Biol. 2017, 435, 62–77. [Google Scholar] [CrossRef] [Green Version]
- Alisafaei, F.; Jokhun, D.S.; Shivashankar, G.; Shenoy, V.B. Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl. Acad. Sci. USA 2019, 116, 13200–13209. [Google Scholar] [CrossRef] [Green Version]
- Fabrikant, G.; Gupta, S.; Shivashankar, G.; Kozlov, M.M. Model of T-cell nuclear deformation by the cortical actin layer. Biophys. J. 2013, 105, 1316–1323. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.G.; Yenepalli, A.; Denais, C.M.; Rape, A.; Beach, J.R.; Wang, Y.-l.; Schiemann, W.P.; Baskaran, H.; Lammerding, J.; Egelhoff, T.T. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell Biol. 2015, 210, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.B.; Whisler, J.A.; Jeon, J.S.; Kamm, R.D. Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integr. Biol. 2013, 5, 1262–1271. [Google Scholar] [CrossRef] [Green Version]
- Shenoy, V.B.; Wang, H.; Wang, X. A chemo-mechanical free-energy-based approach to model durotaxis and extracellular stiffness-dependent contraction and polarization of cells. Interface Focus 2016, 6, 20150067. [Google Scholar] [CrossRef] [Green Version]
- Dahl, K.N.; Kahn, S.M.; Wilson, K.L.; Discher, D.E. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004, 117, 4779–4786. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wen, J.; Xiao, Y.; Liu, J.; Hopyan, S.; Radisic, M.; Simmons, C.A.; Sun, Y. In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS Nano 2014, 8, 3821–3828. [Google Scholar] [CrossRef] [PubMed]
- Funkhouser, C.M.; Sknepnek, R.; Shimi, T.; Goldman, A.E.; Goldman, R.D.; De La Cruz, M.O. Mechanical model of blebbing in nuclear lamin meshworks. Proc. Natl. Acad. Sci. USA 2013, 110, 3248–3253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wren, N.S.; Zhong, Z.; Schwartz, R.S.; Dahl, K.N. Modeling nuclear blebs in a nucleoskeleton of independent filament networks. Cell. Mol. Bioeng. 2012, 5, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, S.; Mathad, S.S.; Sharma, G.; Raju, S.R.; Reddy, U.B.; Das, S.; Ananthasuresh, G. A Nondimensional Model Reveals Alterations in Nuclear Mechanics upon Hepatitis C Virus Replication. Biophys. J. 2019, 116, 1328–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y. A model of cell motility leading to biphasic dependence of transport speed on adhesive strength. J. Mech. Phys. Solids 2010, 58, 502–514. [Google Scholar] [CrossRef] [Green Version]
- Keren, K.; Pincus, Z.; Allen, G.M.; Barnhart, E.L.; Marriott, G.; Mogilner, A.; Theriot, J.A. Mechanism of shape determination in motile cells. Nature 2008, 453, 475. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y. Mechanics model for actin-based motility. Phys. Rev. E 2009, 79, 021916. [Google Scholar] [CrossRef] [Green Version]
- Atilgan, E.; Wirtz, D.; Sun, S.X. Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 2006, 90, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.P.; Richmond, D.L.; Maibaum, L.; Pronk, S.; Geissler, P.L.; Fletcher, D.A. Membrane-induced bundling of actin filaments. Nat. Phys. 2008, 4, 789–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Gong, Z.; Lin, Y.; Chinthapenta, V.; Li, Q.; Webster, T.J.; Sheldon, B.W. Disordered topography mediates filopodial extension and morphology of cells on stiff materials. Adv. Funct. Mater. 2017, 27, 1702689. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, Q.; Qian, J.; Lin, Y.; Shenoy, V.B. Response of biopolymer networks governed by the physical properties of cross-linking molecules. Soft Matter 2016, 12, 2537–2541. [Google Scholar] [CrossRef] [Green Version]
- Broedersz, C.P.; MacKintosh, F.C. Modeling semiflexible polymer networks. Rev. Mod. Phys. 2014, 86, 995. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Wei, X.; Qian, J.; Sze, K.Y.; Shenoy, V.B. A combined finite element-Langevin dynamics (FEM-LD) approach for analyzing the mechanical response of bio-polymer networks. J. Mech. Phys. Solids 2014, 62, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Tosun, A.B.; Guo, J.; Chen, C.; Wang, W.; Ozolek, J.A.; Rohde, G.K. Cancer diagnosis by nuclear morphometry using spatial information. Pattern Recognit. Lett. 2014, 42, 115–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gann, P.H.; Deaton, R.; Amatya, A.; Mohnani, M.; Rueter, E.E.; Yang, Y.; Ananthanarayanan, V. Development of a nuclear morphometric signature for prostate cancer risk in negative biopsies. PLoS ONE 2013, 8, e69457. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, C.; Yao, J.; Xia, X.; Lin, Y. Modelling Nuclear Morphology and Shape Transformation: A Review. Membranes 2021, 11, 540. https://0-doi-org.brum.beds.ac.uk/10.3390/membranes11070540
Fang C, Yao J, Xia X, Lin Y. Modelling Nuclear Morphology and Shape Transformation: A Review. Membranes. 2021; 11(7):540. https://0-doi-org.brum.beds.ac.uk/10.3390/membranes11070540
Chicago/Turabian StyleFang, Chao, Jiaxing Yao, Xingyu Xia, and Yuan Lin. 2021. "Modelling Nuclear Morphology and Shape Transformation: A Review" Membranes 11, no. 7: 540. https://0-doi-org.brum.beds.ac.uk/10.3390/membranes11070540