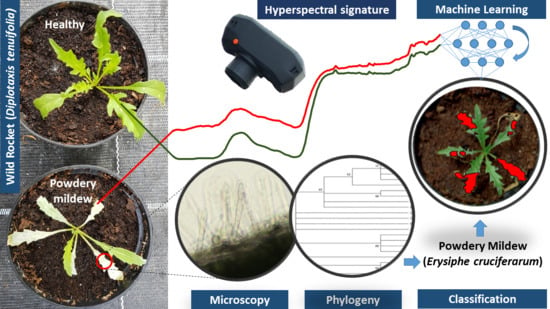
Powdery Mildew Caused by Erysiphe cruciferarum on Wild Rocket (Diplotaxis tenuifolia): Hyperspectral Imaging and Machine Learning Modeling for Non-Destructive Disease Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Greenhouse Experiment
2.2. Pathogen Analysis
2.3. Acquisiton and Preprocessing of Hyperspectral Images
2.4. Variable Pre-Processing and Machine Learning Algorithm
3. Results
3.1. First Report of Powdery Mildew on Wild Rocket
3.2. Hyperspectral Signature of Healthy and Powdery Mildew-Infected Wild Rocket
3.3. Models for Identifying Disease Spots on the Leaves
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Glawe, D.A. The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, M.; Tobin, P.C. Sequencing herbarium specimens of a common detrimental plant disease (powdery mildew). Phytopathology 2020, 110, 7. [Google Scholar] [CrossRef]
- Sambucci, O.; Alston, J.M.; Fuller, K.B.; Lusk, J. The pecuniary and nonpecuniary costs of powdery mildew and the potential value of resistant grape varieties in California. Am. J. Enol. Vitic. 2019, 70, 177–187. [Google Scholar] [CrossRef]
- Jee, H.J.; Shim, C.K.; Ryu, K.Y.; Shin, H.D. Symptoms and damages of powdery mildew on leafy lettuce caused by Podosphaera fusca. Res. Plant Dis. 2006, 12, 294–297. [Google Scholar] [CrossRef] [Green Version]
- Williamson, C.J.; Smith, W.M. Fungicidal control of powdery mildew and its effect on yield, digestibility and chemical composition of eight forage rape cultivars. J. Agric. Sci. 1986, 107, 385–391. [Google Scholar] [CrossRef]
- Asher, M.J.C.; Williams, G.E. Forecasting the national incidence of sugar-beet powdery mildew from weather data in Britain. Plant Pathol. 1991, 40, 100–107. [Google Scholar] [CrossRef]
- Penaud, A. Chemical control and yield losses caused by Erysiphe cruciferarum on oilseed rape in France. In Proceedings of the 10th International Rapeseed Congress, Canberra, Australia, 26–29 September 1999; Wratten, N., Salisbury, P.A., Eds.; The Regional Institute Ltd.: Canberra, Australia, 1999. [Google Scholar]
- Enright, S.M.; Cipollini, D. Infection by powdery mildew Erysiphe cruciferarum (Erysiphaceae) strongly affects growth and fitness of Alliaria petiolata (Brassicaceae). Am. J. Bot. 2007, 94, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, J.; Arumugam, S. Early detection of powdery mildew disease for betelvine plants using digital image analysis. Int. J. Mod. Eng. Res. 2012, 2, 2581–2583. [Google Scholar]
- Mahmud, M.S.; Zaman, Q.U.; Esau, T.J.; Chang, Y.K.; Price, G.W.; Prithiviraj, B. Real-time detection of strawberry powdery mildew disease using a mobile machine vision system. Agronomy 2020, 10, 1027. [Google Scholar] [CrossRef]
- Baudry, J.; Alomar, O.; Begg, G.; Büren, C.; Chenaux, B.; Elmquist, H.; Flamm, C.W.; Gosme, M.; Guerin, O.; Jeanneret, P.; et al. EIP-AGRI Focus Group Benefits of Landscape Features for Arable Crop Production. 2016. Available online: https://hal.inrae.fr/hal-02800942 (accessed on 10 February 2021).
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Behmann, J.; Steinrücken, J.; Plümer, L. Detection of early plant stress responses in hyperspectral images. ISPRS J. Photogramm. Remote Sens. 2014, 93, 98–111. [Google Scholar] [CrossRef]
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Rouphael, Y.; Spíchal, L.; Panzarová, K.; Casa, R.; Colla, G. High-throughput plant phenotyping for developing novel biostimulants: From lab to field or from field to lab? Front. Plant Sci. 2018, 9, 1197. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Plaza, J.; Plaza, A. Deep learning classifiers for hyperspectral imaging: A review. ISPRS J. Photogramm. Remote Sens. 2019, 158, 279–317. [Google Scholar] [CrossRef]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of leaf-scale wheat powdery mildew (Blumeria graminis f. sp. tritici) combining hyperspectral imaging and an SVM classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Roberts, P.; Kakarla, S.C. Detecting powdery mildew disease in squash at different stages using UAV-based hyperspectral imaging and artificial intelligence. Biosyst. Eng. 2020, 197, 135–148. [Google Scholar] [CrossRef]
- Lin, K.; Gong, L.; Huang, Y.; Liu, C.; Pan, J. Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front. Plant Sci. 2019, 10, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Roncal, C.; López-Maestresalas, A.; Lopez-Molina, C.; Jarén, C.; Urrestarazu, J.; Santesteban, L.G.; Arazuri, S. Hyperspectral imaging to assess the presence of powdery mildew (Erysiphe necator) in cv. Carignan noir grapevine bunches. Agronomy 2020, 10, 88. [Google Scholar] [CrossRef] [Green Version]
- Frezza, D.; Logegaray, V.R.; León, A.P.; Harris, M.; Chiesa, A. Rocket (Eruca sativa Mill.) quality affected by preharvest and postharvest factors. Acta Hort. 2010, 875, 357–364. [Google Scholar] [CrossRef]
- Buttaro, D.; Renna, M.; Gerardi, C.; Blando, F.; Santamaria, P.; Serio, F. Soilless production of wild rocket as affected by greenhouse coverage with photovoltaic modules. Acta Sci. Pol. Hortorum Cultus 2016, 15, 129–142. [Google Scholar]
- Bonasia, A.; Lazzizera, C.; Elia, A.; Conversa, G. Nutritional, biophysical and physiological characteristics of wild rocket genotypes as affected by soilless cultivation system, salinity level of nutrient solution and growing period. Front. Plant Sci. 2017, 8, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonasia, A.; Conversa, G.; Lazzizera, C.; Elia, A. Post-harvest performance of ready-to-eat wild rocket salad as affected by growing period, soilless cultivation system and genotype. Postharvest Biol. Technol. 2019, 156, 110909. [Google Scholar] [CrossRef]
- Caruso, G.; Parrella, G.; Giorgini, M.; Nicoletti, R. Crop systems, quality and protection of Diplotaxis tenuifolia. Agriculture 2018, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Attanayake, R.N.; Glawe, D.A.; Dugan, F.M.; Chen, W. Erysiphe trifolii causing powdery mildew of lentil (Lens culinaris). Plant Dis. 2009, 93, 797–803. [Google Scholar] [CrossRef] [Green Version]
- Pastirčáková, K.; Jankovics, T.; Komáromi, J.; Pintye, A.; Pastirčák, M. Genetic diversity and host range of powdery mildews on Papaveraceae. Mycol. Prog. 2016, 15, 36. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Alkooranee, J.T.; Liu, S.; Aledan, T.R.; Yin, Y.; Li, M. First report of powdery mildew caused by Erysiphe cruciferarum on Brassica napus in China. Plant Dis. 2015, 99, 1651. [Google Scholar] [CrossRef]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. R Package Version 3.4-5. 2020. Available online: https://CRAN.R-project.org/package=raster (accessed on 10 February 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 10 February 2021).
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-86. 2020. Available online: https://CRAN.R-project.org/package=caret (accessed on 10 February 2021).
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 3, 1223–1232. [Google Scholar] [CrossRef]
- Purnell, T.J.; Sivanesan, A. Erysiphe cruciferarum. [Descriptions of Fungi and Bacteria]. IMI Descr. Fungi Bact. 1970. [Google Scholar] [CrossRef]
- Choi, J.K.; Kim, J.H.; Cho, S.E.; Shin, H.D. First report of powdery mildew caused by Erysiphe cruciferarum on garden cress in Korea. Plant Dis. 2013, 97, 998. [Google Scholar] [CrossRef]
- Zhao, H.H.; Xing, H.H.; Liang, C.; Yang, X.Y.; Cho, S.E.; Shin, H.D. First report of powdery mildew caused by Erysiphe cruciferarum on Chinese cabbage in China. Plant Dis. 2014, 98, 421. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Glawe, D.A.; McPhee, K.E.; Dugan, F.M.; Chen, W. Erysiphe trifolii—A newly recognized powdery mildew pathogen of pea. Plant Pathol. 2010, 59, 712–720. [Google Scholar] [CrossRef]
- Choi, H.W.; Choi, Y.J.; Kim, D.S.; Hwang, I.S.; Choi, D.S.; Kim, N.H.; Lee, D.H.; Shin, H.D.; Nam, J.; Hwang, B.K. First report of powdery mildew caused by Erysiphe cruciferarum on Arabidopsis thaliana in Korea. Plant Pathol. J. 2009, 25, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Meeboon, J.; Takamatsu, S. Notes on powdery mildews (Erysiphales) in Thailand II. Erysiphe species on Adoxaceae Anacardiaceae, Apocynaceae, Araliaceae, Aristolochiaceae, Bixaceae, Brassicaceae, Cleomaceae, Convolvulaceae, Cucurbitaceae and Euphorbiacea. Trop. Plant Pathol. 2016, 41, 357–369. [Google Scholar] [CrossRef]
- Gunasinghe, N.; You, M.P.; Lanoiselet, V.; Eyres, N.; Barbetti, M.J. First report of powdery mildew caused by Erysiphe cruciferarum on Brassica campestris var. pekinensis, B. carinata, Eruca sativa, E. vesicaria in Australia and on B. rapa and B. oleracea var. capitata in Western Australia. Plant Dis. 2013, 97, 9. [Google Scholar] [CrossRef] [PubMed]
- Betz, E.C.; Punja, Z.K. Management of powdery mildew, caused by Erysiphe cruciferarum, on wasabi (Wasabia japonica) plants in British Columbia. Can. J. Plant Pathol. 2020. [Google Scholar] [CrossRef]
- Nanjundan, J.; Manjunatha, C.; Radhamani, J.; Thakur, A.K.; Yadav, R.; Kumar, A.; Meena, M.L.; Tyagi, R.K.; Yadava, D.K.; Singh, D. Identification of new source of resistance to powdery mildew of Indian mustard and studying its inheritance. Plant Pathol. J. 2020, 36, 111–120. [Google Scholar] [CrossRef]
- Cipollini, D.; Davis, S.; Lieurance, D.; Cipollini, K.; Bahn, V. Biogeographic variation in resistance of the invasive plant, Alliaria petiolata, to a powdery mildew fungus and effect of resistance on competitive dynamics. Biol. Invasions 2020, 22, 1657–1668. [Google Scholar] [CrossRef] [Green Version]
- Bahçecioǧlu, Z.; Kabaktepe, S.; Yildiz, B. Microfungi isolated from plants in Kahramanmarafl Province, Turkey. Turk. J. Bot. 2006, 30, 419–434. [Google Scholar]
- Nicoletti, R.; Raimo, F.; Miccio, G. Diplotaxis tenuifolia: Biology, production and properties. Eur. J. Plant Sci. Biotechnol. 2007, 1, 36–43. [Google Scholar]
- Desai, A.G.; Chattopadhyay, C.; Agrawal, R.; Kumar, A.; Meena, R.L.; Meena, P.D.; Sharma, K.C.; Srinivasa Rao, M.; Prasad, Y.G.; Ramakrishna, Y.S. Brassica juncea powdery mildew epidemiology and weather based forecasting models for India—A case study. J. Plant Dis. Prot. 2004, 5, 429–438. [Google Scholar]
- Garibaldi, A.; Bertetti, D.; Gullino, M.L. Outbreak of powdery mildew caused by Erysiphe cruciferarum on spider flower (Cleome hassleriana) in Italy. Plant Dis. 2009, 93, 963. [Google Scholar] [CrossRef]
- Vellios, E.; Karkanis, A.; Bilalis, D. Powdery mildew (Erysiphe cruciferarum) infection on camelina (Camelina sativa) under Mediterranean conditions and the role of wild mustard (Sinapis arvensis) as alternative host of this pathogen. Emir. J. Food Agric. 2017, 29, 639–642. [Google Scholar] [CrossRef] [Green Version]
- Uloth, M.B.; You, M.P.; Barbetti, M.J. Plant age and ambient temperature: Significant drivers for powdery mildew (Erysiphe cruciferarum) epidemics on oilseed rape (Brassica napus). Plant Pathol. 2018, 67, 445–456. [Google Scholar] [CrossRef]
- Adam, L.; Somerville, S.C. Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 1996, 9, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Paul, N.D.; Ayres, P.G. The impact of a pathogen (Puccinia lagenophorae) on populations of groundsel (Senecio vulgaris) overwintering in the field: I. mortality, vegetative growth and the development of size hierarchies. J. Ecol. 1986, 74, 1069–1084. [Google Scholar] [CrossRef]
- Kuckenberg, J.; Tartachnyk, I.; Schmitz-Eiberger, M.; Noga, G. Early detection of leaf rust and powdery mildew infections on wheat leaves by PAM fluorescence imaging. In Proceedings of the 6th European Conference on Precision Agriculture, Skiathos, Greece, 3–6 June 2007; Stafford, J.V., Ed.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2007; pp. 515–521. [Google Scholar]
- Mahlein, A.K.; Steiner, U.; Hillnhütter, C.; Dehne, H.W.; Oerke, E.C. Hyperspectral imaging for small-scale analysis of symptoms caused by different sugar beet diseases. Plant Methods 2012, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Tartachnyk, I.; Rademacher, I.; Kühbauch, W. Distinguishing nitrogen deficiency and fungal infection of winter wheat by laser-induced fluorescence. Precis. Agric. 2006, 7, 281–293. [Google Scholar] [CrossRef]
- Devadas, R.; Lamb, D.W.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precis. Agric. 2009, 10, 459–470. [Google Scholar] [CrossRef]
- Abu-Foul, S.; Raskin, V.I.; Sztejnberg, A.; Marder, J.B. Disruption of chlorophyll organization and function in powdery mildew-diseased cucumber leaves and its control by the hyperparasite Ampelomyces quisqualis. Phytopathology 1996, 86, 195–199. [Google Scholar] [CrossRef]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral imaging for presymptomatic detection of tobacco disease with successive projections algorithm and machine-learning classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogewoning, S.W.; Trouwborst, G.; Maljaars, H.; Poorter, H.; van Ieperen, W.; Harbinson, J. Blue light dose–responses of leaf photosynthesis, morphology, and chemical composition of Cucumis sativus grown under different combinations of red and blue light. J. Exp. Bot. 2010, 61, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, W.; Tong, Y.; Yang, Q. Leaf morphology, photosynthetic performance, chlorophyll fluorescence, stomatal development of lettuce (Lactuca sativa L.) exposed to different ratios of red light to blue light. Front. Plant Sci. 2016, 7, 250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calderón, R.; Navas-Cortés, J.A.; Lucena, C.; Zarco-Tejada, P.J. High-resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band spectral indices. Remote Sens. Environ. 2013, 139, 231–245. [Google Scholar] [CrossRef]
- Hniličková, H.; Hnilička, F.; Martinkova, J.; Kraus, K. Effects of salt stress on water status, photosynthesis and chlorophyll fluorescence of rocket. Plant Soil Environ. 2017, 63, 362–367. [Google Scholar]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pane, C.; Manganiello, G.; Nicastro, N.; Cardi, T.; Carotenuto, F. Powdery Mildew Caused by Erysiphe cruciferarum on Wild Rocket (Diplotaxis tenuifolia): Hyperspectral Imaging and Machine Learning Modeling for Non-Destructive Disease Detection. Agriculture 2021, 11, 337. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11040337
Pane C, Manganiello G, Nicastro N, Cardi T, Carotenuto F. Powdery Mildew Caused by Erysiphe cruciferarum on Wild Rocket (Diplotaxis tenuifolia): Hyperspectral Imaging and Machine Learning Modeling for Non-Destructive Disease Detection. Agriculture. 2021; 11(4):337. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11040337
Chicago/Turabian StylePane, Catello, Gelsomina Manganiello, Nicola Nicastro, Teodoro Cardi, and Francesco Carotenuto. 2021. "Powdery Mildew Caused by Erysiphe cruciferarum on Wild Rocket (Diplotaxis tenuifolia): Hyperspectral Imaging and Machine Learning Modeling for Non-Destructive Disease Detection" Agriculture 11, no. 4: 337. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11040337