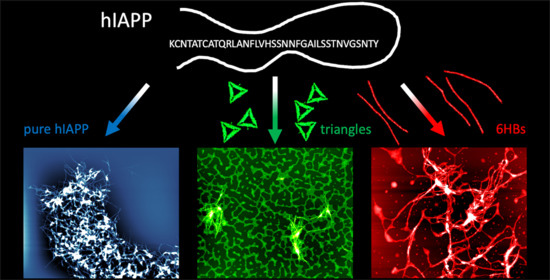
Effect of DNA Origami Nanostructures on hIAPP Aggregation
Abstract
:1. Introduction
2. Materials and Methods
2.1. hIAPP Preparation
2.2. DNA Origami and Genomic dsDNA Preparation
2.3. Sample Preparation
2.4. Turbidity Measurements
2.5. AFM
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Törnquist, M.; Michaels, T.C.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.; Knowles, T.P.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [Green Version]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid–from bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef]
- Otzen, D.; Riek, R. Functional amyloids. Cold Spring Harbor. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.D.; Cohen, A.S. Review: History of the Amyloid Fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höppener, J.W.M.; Ahrén, B.; Lips, C.J.M. Islet Amyloid and Type 2 Diabetes Mellitus. N. Engl. J. Med. 2000, 343, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Khemtémourian, L.; Antoinette Killian, J.; Hö∂ppener, J.W.; Engel, M.F. Recent Insights in Islet Amyloid Polypeptide-Induced Membrane Disruption and Its Role in Cell Death in Type 2 Diabetes Mellitus. Exp. Diabetes Res. 2008, 2008. [Google Scholar] [CrossRef] [Green Version]
- Eichner, T.; Radford, S.E. A Diversity of Assembly Mechanisms of a Generic Amyloid Fold. Mol. Cell 2011, 43, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Keller, A.; Grundmeier, G. Amyloid aggregation at solid-liquid interfaces: Perspectives of studies using model surfaces. Appl. Surf. Sci. 2020, 506, 144991. [Google Scholar] [CrossRef]
- Härd, T.; Lendel, C. Inhibition of Amyloid Formation. J. Mol. Biol. 2012, 421, 441–465. [Google Scholar] [CrossRef]
- Owen, M.C.; Gnutt, D.; Gao, M.; Wärmländer, S.K.T.S.; Jarvet, J.; Gräslund, A.; Winter, R.; Ebbinghaus, S.; Strodel, B. Effects of in vivo conditions on amyloid aggregation. Chem. Soc. Rev. 2019, 48, 3946–3996. [Google Scholar] [CrossRef]
- Bao, G.; Mitragotri, S.; Tong, S. Multifunctional Nanoparticles for Drug Delivery and Molecular Imaging. Ann. Rev. Biomed. Eng. 2013, 15, 253–282. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, M.P.; Åberg, C.; Salvati, A.; Dawson, K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012, 7, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Kalhor, H.R.; Laurent, S.; Lynch, I. Protein fibrillation and nanoparticle interactions: Opportunities and challenges. Nanoscale 2013, 5, 2570–2588. [Google Scholar] [CrossRef]
- John, T.; Gladytz, A.; Kubeil, C.; Martin, L.L.; Risselada, H.J.; Abel, B. Impact of nanoparticles on amyloid peptide and protein aggregation: A review with a focus on gold nanoparticles. Nanoscale 2018, 10, 20894–20913. [Google Scholar] [CrossRef]
- Ke, P.C.; Pilkington, E.H.; Sun, Y.; Javed, I.; Kakinen, A.; Peng, G.; Ding, F.; Davis, T.P. Mitigation of Amyloidosis with Nanomaterials. Adv. Mater. 2020, 32, 1901690. [Google Scholar] [CrossRef]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. 2020, 59, 15818–15833. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Liu, S.; Liu, J.; Wang, Z.-G.; Ding, B. Rationally Designed DNA-Origami Nanomaterials for Drug Delivery In Vivo. Adv. Mater. 2019, 31, 1804785. [Google Scholar] [CrossRef]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.-Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Wiraja, C.; Zhu, Y.; Lio, D.C.S.; Yeo, D.C.; Xie, M.; Fang, W.; Li, Q.; Zheng, M.; Van Steensel, M.; Wang, L.; et al. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat. Commun. 2019, 10, 1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Song, L.; Liu, S.; Jiang, Q.; Liu, Q.; Li, N.; Wang, Z.-G.; Ding, B. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018, 18, 3328–3334. [Google Scholar] [CrossRef] [PubMed]
- Palazzolo, S.; Hadla, M.; Spena, C.R.; Bayda, S.; Kumar, V.; Lo Re, F.; Adeel, M.; Caligiuri, I.; Romano, F.; Corona, G.; et al. Proof-of-Concept Multistage Biomimetic Liposomal DNA Origami Nanosystem for the Remote Loading of Doxorubicin. ACS Med. Chem. Lett. 2019, 10, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Ge, Z.; Im, H.-J.; England, C.G.; Ni, D.; Hou, J.; Zhang, L.; Kutyreff, C.J.; Yan, Y.; Liu, Y.; et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2018, 2, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, D.; Rosenkrans, Z.T.; Barnhart, T.E.; Ehlerding, E.B.; Ni, D.; Engle, J.W.; Cai, W. Aptamer-Conjugated Framework Nucleic Acids for the Repair of Cerebral Ischemia-Reperfusion Injury. Nano Lett. 2019, 19, 7334–7341. [Google Scholar] [CrossRef]
- Hajiraissi, R.; Hanke, M.; Yang, Y.; Duderija, B.; Gonzalez Orive, A.; Grundmeier, G.; Keller, A. Adsorption and Fibrillization of Islet Amyloid Polypeptide at Self-Assembled Monolayers Studied by QCM-D, AFM, and PM-IRRAS. Langmuir 2018, 34, 3517–3524. [Google Scholar] [CrossRef]
- Bui, H.; Onodera, C.; Kidwell, C.; Tan, Y.; Graugnard, E.; Kuang, W.; Lee, J.; Knowlton, W.B.; Yurke, B.; Hughes, W.L. Programmable Periodicity of Quantum Dot Arrays with DNA Origami Nanotubes. Nano Lett. 2010, 10, 3367–3372. [Google Scholar] [CrossRef]
- Opherden, L.; Oertel, J.; Barkleit, A.; Fahmy, K.; Keller, A. Paramagnetic Decoration of DNA Origami Nanostructures by Eu3+ Coordination. Langmuir 2014, 30, 8152–8159. [Google Scholar] [CrossRef]
- Kollmann, F.; Ramakrishnan, S.; Shen, B.; Grundmeier, G.; Kostiainen, M.A.; Linko, V.; Keller, A. Superstructure-Dependent Loading of DNA Origami Nanostructures with a Groove-Binding Drug. ACS Omega 2018, 3, 9441–9448. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Brender, J.R.; Krishnamoorthy, J.; Sciacca, M.F.M.; Vivekanandan, S.; D’Urso, L.; Chen, J.; La Rosa, C.; Ramamoorthy, A. Probing the Sources of the Apparent Irreproducibility of Amyloid Formation: Drastic Changes in Kinetics and a Switch in Mechanism Due to Micellelike Oligomer Formation at Critical Concentrations of IAPP. J. Phys. Chem. B 2015, 119, 2886–2896. [Google Scholar] [CrossRef]
- Broersen, K.; Jonckheere, W.; Rozenski, J.; Vandersteen, A.; Pauwels, K.; Pastore, A.; Rousseau, F.; Schymkowitz, J. A standardized and biocompatible preparation of aggregate-free amyloid beta peptide for biophysical and biological studies of Alzheimer’s disease. Protein Eng. Des. Sel. 2011, 24, 743–750. [Google Scholar] [CrossRef] [Green Version]
- Stine, W.B.; Jungbauer, L.; Yu, C.; LaDu, M.J. Preparing synthetic Aβ in different aggregation states. Methods Mol. Biol. 2011, 670, 13–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lim, H.J.; Son, A. Characterization of denaturation and renaturation of DNA for DNA hybridization. Environ. Anal. Health Toxicol. 2014, 29, e2014007. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Ijäs, H.; Linko, V.; Keller, A. Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 2018, 16, 342–349. [Google Scholar] [CrossRef]
- Xue, C.; Lin, T.Y.; Chang, D.; Guo, Z. Thioflavin T as an amyloid dye: Fibril quantification, optimal concentration and effect on aggregation. R. Soc. Open Sci. 2017, 4, 160696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biancalana, M.; Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murudkar, S.; Mora, A.K.; Jakka, S.; Singh, P.K.; Nath, S. Ultrafast molecular rotor based DNA sensor: An insight into the mode of interaction. J. Photochem. Photobiol. A Chem. 2014, 295, 17–25. [Google Scholar] [CrossRef]
- Murudkar, S.; Mora, A.K.; Singh, P.K.; Nath, S. Ultrafast molecular rotor: An efficient sensor for premelting of natural DNA. Chem. Commun. 2012, 48, 5301–5303. [Google Scholar] [CrossRef]
- Zhou, W.; Yu, Z.; Ma, G.; Jin, T.; Li, Y.; Fan, L.; Li, X. Thioflavin T specifically brightening “Guanine Island” in duplex-DNA: A novel fluorescent probe for single-nucleotide mutation. Analyst 2019, 144, 2284–2290. [Google Scholar] [CrossRef]
- Come, J.H.; Fraser, P.E.; Lansbury, P.T. A kinetic model for amyloid formation in the prion diseases: Importance of seeding. Proc. Natl. Acad. Sci. USA 1993, 90, 5959–5963. [Google Scholar] [CrossRef] [Green Version]
- Jarrett, J.T.; Berger, E.P.; Lansbury, P.T. The carboxy terminus of the. beta. amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry 1993, 32, 4693–4697. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.P.; Martí, A.A. Facile Methodology for Monitoring Amyloid-β Fibrillization. ACS Chem. Neurosci. 2012, 3, 896–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, A.; Li, Z.; Yu, L.; Wang, H.; Wang, E. Plasmid DNA Network on a Mica Substrate Investigated by Atomic Force Microscopy. Anal. Sci. 2001, 17, 583–584. [Google Scholar] [CrossRef] [Green Version]
- Bezanilla, M.; Manne, S.; Laney, D.E.; Lyubchenko, Y.L.; Hansma, H.G. Adsorption of DNA to Mica, Silylated Mica, and Minerals: Characterization by Atomic Force Microscopy. Langmuir 1995, 11, 655–659. [Google Scholar] [CrossRef]
- Cai, L.; Tabata, H.; Kawai, T. Self-assembled DNA networks and their electrical conductivity. Appl. Phys. Lett. 2000, 77, 3105–3106. [Google Scholar] [CrossRef]
- Wu, A.; Li, Z.; Zhou, H.; Zheng, J.; Wang, E. Construction and control of plasmid DNA network. Analyst 2002, 127, 585–587. [Google Scholar] [CrossRef]
- Kanno, T.; Tanaka, H.; Miyoshi, N.; Kawai, T. Formation and control of two-dimensional deoxyribonucleic acid network. Appl. Phys. Lett. 2000, 77, 3848–3850. [Google Scholar] [CrossRef]
- Xiao, Z. AFM observations of self-assembled lambda DNA network on silanized mica. Thin Solid Films 2003, 438–439, 114. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, D.; Zhang, Y.; Xu, F.; Li, Z. DNA adsorption and desorption on mica surface studied by atomic force microscopy. Appl. Surf. Sci. 2011, 257, 6560–6567. [Google Scholar] [CrossRef]
- Murayama, H.; Yoshikawa, K. Thermodynamics of the Collapsing Phase Transition in a Single Duplex DNA Molecule. J. Phys. Chem. B 1999, 103, 10517–10523. [Google Scholar] [CrossRef]
- Hirota, N.; Edskes, H.; Hall, D. Unified theoretical description of the kinetics of protein aggregation. Biophys. Rev. 2019, 11, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro-Lago, C.; Quinlan-Pluck, F.; Lynch, I.; Dawson, K.A.; Linse, S. Dual Effect of Amino Modified Polystyrene Nanoparticles on Amyloid β Protein Fibrillation. ACS Chem. Neurosci. 2010, 1, 279–287. [Google Scholar] [CrossRef]
- Wang, W.; Han, Y.; Fan, Y.; Wang, Y. Effects of Gold Nanospheres and Nanocubes on Amyloid-β Peptide Fibrillation. Langmuir 2019, 35, 2334–2342. [Google Scholar] [CrossRef]
- Estrich, N.A.; Hernandez-Garcia, A.; de Vries, R.; LaBean, T.H. Engineered Diblock Polypeptides Improve DNA and Gold Solubility during Molecular Assembly. ACS Nano 2017, 11, 831–842. [Google Scholar] [CrossRef]
- Agarwal, N.P.; Matthies, M.; Gür, F.N.; Osada, K.; Schmidt, T.L. Block Copolymer Micellization as a Protection Strategy for DNA Origami. Angew. Chem. Int. Ed. 2017, 56, 5460–5464. [Google Scholar] [CrossRef]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef]
- Kiviaho, J.K.; Linko, V.; Ora, A.; Tiainen, T.; Järvihaavisto, E.; Mikkilä, J.; Tenhu, H.; Nonappa; Kostiainen, M.A. Cationic polymers for DNA origami coating—Examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale 2016, 8, 11674–11680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadi, Y.; De Llano, E.; Barišić, I. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale 2018, 10, 7494–7504. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Fang, S.; Zhuang, Y.; Wu, S.; Pan, Q.; Li, L.; Wang, X.; Sun, X.; Liu, B.; Wu, Y. Cationic Albumin Encapsulated DNA Origami for Enhanced Cellular Transfection and Stability. Materials 2019, 12, 949. [Google Scholar] [CrossRef] [Green Version]
- Auvinen, H.; Zhang, H.; Nonappa; Kopilow, A.; Niemelä, E.H.; Nummelin, S.; Correia, A.; Santos, H.A.; Linko, V.; Kostiainen, M.A. Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv. Healthc. Mater. 2017, 6, 1700692. [Google Scholar] [CrossRef] [Green Version]
- Mikkilä, J.; Eskelinen, A.-P.; Niemelä, E.H.; Linko, V.; Frilander, M.J.; Törmä, P.; Kostiainen, M.A. Virus-Encapsulated DNA Origami Nanostructures for Cellular Delivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Meyer, T.A.; Modlin, C.; Zuo, X.; Conticello, V.P.; Ke, Y. Structurally Ordered Nanowire Formation from Co-Assembly of DNA Origami and Collagen-Mimetic Peptides. J. Am. Chem. Soc. 2017, 139, 14025–14028. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.; Strehle, D.; Käs, J. Counterion-induced formation of regular actin bundle networks. Soft Matter 2012, 8, 931–936. [Google Scholar] [CrossRef]
- Huber, F.; Strehle, D.; Schnauß, J.; Käs, J. Formation of regularly spaced networks as a general feature of actin bundle condensation by entropic forces. New J. Phys. 2015, 17, 043029. [Google Scholar] [CrossRef] [Green Version]
- Glaser, M.; Schnauß, J.; Tschirner, T.; Schmidt, B.U.S.; Moebius-Winkler, M.; Käs, J.A.; Smith, D.M. Self-assembly of hierarchically ordered structures in DNA nanotube systems. New J. Phys. 2016, 18, 055001. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, S.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Keller, A.; Linko, V. Real-Time Observation of Superstructure-Dependent DNA Origami Digestion by DNase I Using High-Speed Atomic Force Microscopy. ChemBioChem 2019, 20, 2818–2823. [Google Scholar] [CrossRef]
- Suma, A.; Stopar, A.; Nicholson, A.W.; Castronovo, M.; Carnevale, V. Global and local mechanical properties control endonuclease reactivity of a DNA origami nanostructure. Nucl. Acids Res. 2020, 48, 4672–4680. [Google Scholar] [CrossRef] [Green Version]
- Stopar, A.; Coral, L.; Di Giacomo, S.; Adedeji, A.F.; Castronovo, M. Binary control of enzymatic cleavage of DNA origami by structural antideterminants. Nucl. Acids Res. 2017, 46, 995–1006. [Google Scholar] [CrossRef] [Green Version]
- Julin, S.; Korpi, A.; Nonappa; Shen, B.; Liljeström, V.; Ikkala, O.; Keller, A.; Linko, V.; Kostiainen, M.A. DNA origami directed 3D nanoparticle superlattice via electrostatic assembly. Nanoscale 2019, 11, 4546–4551. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Okahata, Y. A DNA−Lipid Complex in Organic Media and Formation of an Aligned Cast Film1. J. Am. Chem. Soc. 1996, 118, 10679–10683. [Google Scholar] [CrossRef]
- Manning, G.S. The Persistence Length of DNA Is Reached from the Persistence Length of Its Null Isomer through an Internal Electrostatic Stretching Force. Biophys. J. 2006, 91, 3607–3616. [Google Scholar] [CrossRef] [Green Version]
- Siavashpouri, M.; Wachauf, C.H.; Zakhary, M.J.; Praetorius, F.; Dietz, H.; Dogic, Z. Molecular engineering of chiral colloidal liquid crystals using DNA origami. Nat. Mater. 2017, 16, 849–856. [Google Scholar] [CrossRef]
- Köster, D.V.; Husain, K.; Iljazi, E.; Bhat, A.; Bieling, P.; Mullins, R.D.; Rao, M.; Mayor, S. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc. Natl. Acad. Sci. USA 2016, 113, E1645. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.; Ziebert, F.; Humphrey, D.; Duggan, C.; Steinbeck, M.; Zimmermann, W.; Käs, J. Molecular Motor-Induced Instabilities and Cross Linkers Determine Biopolymer Organization. Biophys. J. 2007, 93, 4445–4452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndlec, F.J.; Surrey, T.; Maggs, A.C.; Leibler, S. Self-organization of microtubules and motors. Nature 1997, 389, 305–308. [Google Scholar] [CrossRef]
- Surrey, T.; Nédélec, F.; Leibler, S.; Karsenti, E. Physical Properties Determining Self-Organization of Motors and Microtubules. Science 2001, 292, 1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibaud, T.; Barry, E.; Zakhary, M.J.; Henglin, M.; Ward, A.; Yang, Y.; Berciu, C.; Oldenbourg, R.; Hagan, M.F.; Nicastro, D.; et al. Reconfigurable self-assembly through chiral control of interfacial tension. Nature 2012, 481, 348–351. [Google Scholar] [CrossRef]
- Lew, D.; Parker, S.E.; Latimer, T.; Abai, A.M.; Kuwahara-Rundell, A.; Doh, S.C.; Yang, Z.-Y.; Laface, D.; Gromkowski, S.H.; Nabel, G.J.; et al. Cancer Gene Therapy Using Plasmid DNA: Pharmacokinetic Study of DNA Following Injection in Mice. Hum. Gene Ther. 1995, 6, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33–37. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanke, M.; Gonzalez Orive, A.; Grundmeier, G.; Keller, A. Effect of DNA Origami Nanostructures on hIAPP Aggregation. Nanomaterials 2020, 10, 2200. https://0-doi-org.brum.beds.ac.uk/10.3390/nano10112200
Hanke M, Gonzalez Orive A, Grundmeier G, Keller A. Effect of DNA Origami Nanostructures on hIAPP Aggregation. Nanomaterials. 2020; 10(11):2200. https://0-doi-org.brum.beds.ac.uk/10.3390/nano10112200
Chicago/Turabian StyleHanke, Marcel, Alejandro Gonzalez Orive, Guido Grundmeier, and Adrian Keller. 2020. "Effect of DNA Origami Nanostructures on hIAPP Aggregation" Nanomaterials 10, no. 11: 2200. https://0-doi-org.brum.beds.ac.uk/10.3390/nano10112200