Integrin-Targeting Dye-Doped PEG-Shell/Silica-Core Nanoparticles Mimicking the Proapoptotic Smac/DIABLO Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. General Methods
2.1.2. c[Arg–Gly–Asp–D-Phe–Lys(hex–5–ynamide)] (cRGD–alkyne)
2.1.3. H–Ala–Val–Pro–Ile–Gly–pent-4-yn-1-amine (AVPI–alkyne)
2.1.4. PEG-Shell/Silica-Core Azide–NPs (NP–N3)
2.1.5. NP–N3 Functionalization with cRGD–alkyne and/or AVPI–alkyne
2.2. Biological Methods
2.2.1. Cells and Culture Conditions
2.2.2. Cell Viability Assays
2.2.3. Apoptosis
2.2.4. Cell Internalization
2.2.5. Competition Experiments
3. Results
3.1. Chemistry
3.2. Cytotoxicity of Peptide–NPs
3.3. Caspase-9 Activity
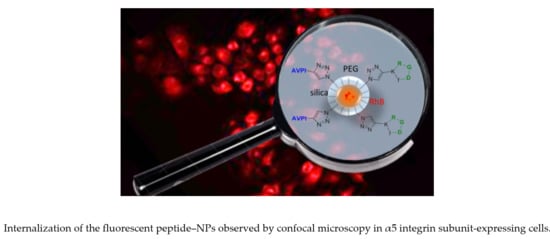
3.4. Cellular Uptake of Peptide–NPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Crook, N.E.; Clem, R.J.; Miller, L.K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993, 67, 2168–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocab, A.J.; Duckett, C.S. Inhibitor of apoptosis proteins as intracellular signalling intermediates. FEBS J. 2016, 283, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Berthelet, J.; Dubrez, L. Regulation of apoptosis by inhibitors of apoptosis (IAPs). Cells 2013, 2, 163–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckelman, B.P.; Drag, M.; Snipas, S.J.; Salvesen, G.S. The mechanism of peptide-binding of IAP BIR domains. Cell Death Differ. 2008, 15, 920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deveraux, Q.L.; Takahashi, R.; Salvesen, G.S.; Reed, J.C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 1997, 388, 300–304. [Google Scholar] [CrossRef]
- Cossu, F.; Milani, M.; Mastrangelo, E.; Lecis, D. Targeting the BIR domains of inhibitor of apoptosis (IAP) proteins in cancer treatment. Comput. Struct. Biotechnol. J. 2019, 17, 142–150. [Google Scholar] [CrossRef]
- Mohamed, M.S.; Bishr, M.K.; Almutairi, F.M.; Ali, A.G. Inhibitors of apoptosis: Clinical implications in cancer. Apoptosis 2017, 22, 1487–1509. [Google Scholar] [CrossRef]
- Rathore, R.; McCallum, J.E.; Varghese, E.; Florea, A.M.; Büsselber, D. Overcoming chemotherapy drug resistance by targeting inhibitors of apoptosis proteins (IAPs). Apoptosis 2017, 22, 898–919. [Google Scholar] [CrossRef]
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Chai, J.; Suber, T.L.; Wu, J.W.; Du, C.; Wang, X.; Shi, Y. Structural basis of IAP recognition by Smac/DIABLO. Nature 2000, 408, 1008–1012. [Google Scholar] [CrossRef]
- Gentilucci, L.; Tolomelli, A.; Squassabia, F. Peptides and peptidomimetics in medicine, surgery and biotechnology. Curr. Med. Chem. 2006, 13, 2449–2466. [Google Scholar] [CrossRef] [PubMed]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical modifications designed to improve peptide stability: Incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mashima, T.; Sato, S.; Mochizuki, M.; Sakamoto, H.; Yamori, T.; Oh-Hara, T.; Tsuruo, T. Predominant suppression of apoptosome by inhibitor of apoptosis protein in non-small cell lung cancer H460 cells: Therapeutic effect of a novel polyarginine conjugated Smac peptide. Cancer Res. 2003, 63, 831–837. [Google Scholar] [PubMed]
- Hossbach, J.; Michalsky, E.; Henklein, P.; Jaeger, M.; Daniel, P.T.; Preissner, R. Inhibiting the inhibitors: Retro-inverso Smac peptides. Peptides 2009, 30, 2374–2379. [Google Scholar] [CrossRef] [PubMed]
- Ardecky, R.J.; Welsh, K.; Finlay, D.; Lee, P.S.; González-López, M.; Ganji, S.R.; Ravanan, P.; Mace, P.D.; Riedl, S.J.; Vuori, K.; et al. Design, synthesis and evaluation of inhibitor of apoptosis protein (IAP) antagonists that are highly selective for the BIR2 domain of XIAP. Bioorg. Med. Chem. Lett. 2013, 23, 4253–4257. [Google Scholar] [CrossRef] [Green Version]
- Bourguet, C.B.; Boulay, P.L.; Claing, A.; Lubell, W.D. Design and synthesis of novel azapeptide activators of apoptosis mediated by caspase-9 in cancer cells. Bioorg. Med. Chem. Lett. 2014, 24, 3361–3365. [Google Scholar] [CrossRef] [PubMed]
- Dadwal, A.; Baldi, A.; Kumar Narang, R. Nanoparticles as carriers for drug delivery in cancer. Artif. Cell Nanomed. Biotechnol. 2018, 46, 295–305. [Google Scholar] [CrossRef]
- Wang, A.Z.; Langer, R.; Farokhzad, O.C. Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 2012, 63, 185–198. [Google Scholar] [CrossRef]
- Kratochvílová, I.; Šebera, J.; Ashcheulov, P.; Golan, M.; Ledvina, M.; Míčová, J.; Mravec, F.; Kovalenko, A.; Zverev, D.; Yavkin, B.; et al. Magnetical and optical properties of nanodiamonds can be tuned by particles surface chemistry: Theoretical and experimental study. J. Phys. Chem. C 2014, 43, 25245–25252. [Google Scholar]
- Jeong, W.-J.; Bu, J.; Kubiatowicz, L.J.; Chen, S.S.; Kim, Y.; Hong, S. Peptide–nanoparticle conjugates: A next generation of diagnostic and therapeutic platforms? Nano Converg. 2018, 5, 38. [Google Scholar] [CrossRef]
- Spicer, C.D.; Jumeaux, C.; Gupta, B.; Stevens, M.M. Peptide and protein nanoparticle conjugates: Versatile platforms for biomedical applications. Chem. Soc. Rev. 2018, 47, 3574–3620. [Google Scholar] [CrossRef] [Green Version]
- Pudlarz, A.; Szemraj, J. Nanoparticles as carriers of proteins, peptides and other therapeutic molecules. Open Life Sci. 2018, 13, 285–298. [Google Scholar] [CrossRef]
- Seneci, P.; Rizzi, M.; Ballabio, L.; Lecis, D.; Conti, A.; Carrara, C.; Licandro, E. SPION-Smac mimetic nano-conjugates: Putative pro-apoptotic agents in oncology. Bioorg. Med. Chem. Lett. 2014, 24, 2374–2378. [Google Scholar] [CrossRef]
- Li, M.; Liu, P.; Gao, G.; Deng, J.; Pan, Z.; Wu, X.; Xie, G.; Yue, C.; Cho, C.H.; Ma, Y.; et al. Smac therapeutic peptide nanoparticles inducing apoptosis of cancer cells for combination chemotherapy with Doxorubicin. ACS Appl. Mater. Interfaces 2015, 7, 8005–8012. [Google Scholar] [CrossRef]
- Arosio, D.; Manzoni, L.; Corno, C.; Perego, P. Integrin-targeted peptide- and peptidomimetic-drug conjugates for the treatment of tumors. Recent Pat. Anti-Cancer 2017, 12, 148–168. [Google Scholar] [CrossRef]
- Borbély, A.; Figueras, E.; Martins, A.; Bodero, L.; Raposo Moreira Dias, A.; López Rivas, P.; Pina, A.; Arosio, D.; Gallinari, P.; Frese, M.; et al. Conjugates of cryptophycin and RGD or isoDGR peptidomimetics for targeted drug delivery. Chem. Open 2019, 8, 737–742. [Google Scholar]
- Conde, J.; Tian, F.; Hernández, Y.; Bao, C.; Cui, D.; Janssen, K.P.; Ibarra, M.R.; Baptista, P.V.; Stoeger, T.; de la Fuente, J.M. In vivo tumor targeting via nanoparticle-mediated therapeutic siRNA coupled to inflammatory response in lung cancer mouse models. Biomaterials 2013, 34, 7744–7753. [Google Scholar] [CrossRef]
- Duro-Castano, A.; Gallon, E.; Decker, C.; Vicent, M.J. Modulating angiogenesis with integrin-targeted nanomedicines. Adv. Drug Deliv. Rev. 2017, 119, 101–119. [Google Scholar] [CrossRef]
- Duret, D.; Grassin, A.; Henry, M.; Jacquet, T.; Thoreau, F.; Denis-Quanquin, S.; Coll, J.L.; Boturyn, D.; Favier, A.; Charreyre, M.T. Polymultivalent polymer-peptide cluster conjugates for an enhanced targeting of cells expressing αvβ3 integrins. Bioconjugate Chem. 2017, 28, 2241–2245. [Google Scholar] [CrossRef]
- Mas-Moruno, C.; Fraioli, R.; Rechenmacher, F.; Neubauer, S.; Kapp, T.G.; Kessler, H. αvβ3- or α5β1-Integrin-selective peptidomimetics for surface coating. Angew. Chem. Int. Ed. 2016, 55, 7048–7067. [Google Scholar] [CrossRef]
- Greco, A.; Maggini, L.; De Cola, L.; De Marco, R.; Gentilucci, L. Diagnostic implementation of fast and selective integrin-mediated adhesion of cancer cells on functionalized Zeolite L Monolayers. Bioconjugate Chem. 2015, 26, 1873–1878. [Google Scholar] [CrossRef]
- De Marco, R.; Greco, A.; Calonghi, N.; Dattoli, S.D.; Baiula, M.; Spampinato, S.; Picchetti, P.; De Cola, L.; Anselmi, M.; Cipriani, F.; et al. Selective detection of alfa4beta1 integrin (VLA-4)-expressing cells using peptide-functionalized nanostructured materials mimicking endothelial surfaces adjacent to inflammatory sites. Pept. Sci. 2018, 110, e23081. [Google Scholar] [CrossRef]
- Nieberler, M.; Reuning, U.; Reichart, F.; Notni, J.; Wester, H.J.; Schwaiger, M.; Weinmüller, M.; Räder, A.; Steiger, K.; Kessler, H. Exploring the Role of RGD-Recognizing Integrins in Cancer. Cancers 2017, 9, 116. [Google Scholar] [CrossRef]
- Katsamakas, S.; Chatzisideri, T.; Thysiadis, S.; Sarli, V. RGD-mediated delivery of small-molecule drugs. Future Med. Chem. 2017, 9, 579–604. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Mingozzi, M.; Manzoni, L.; Arosio, D.; Dal Corso, A.; Manzotti, M.; Innamorati, F.; Pignataro, L.; Lecis, D.; Delia, D.; Seneci, P.; et al. Synthesis and biological evaluation of dual action cyclo-RGD/SMAC mimetic conjugates targeting αvβ3/αvβ5 integrins and IAP proteins. Org. Biomol. Chem. 2014, 12, 3288–3302. [Google Scholar] [CrossRef] [Green Version]
- Swain, J.; Mishra, A.K. Nile red fluorescence for quantitative monitoring of micropolarity and microviscosity of pluronic F127 in aqueous media. Photochem. Photobiol. Sci. 2016, 15, 1400–1407. [Google Scholar] [CrossRef]
- Basak, R.; Bandyopadhyay, R. Encapsulation of hydrophobic drugs in Pluronic F127 micelles: Effects of drug hydrophobicity, solution temperature, and pH. Langmuir 2013, 29, 4350–4356. [Google Scholar] [CrossRef] [Green Version]
- Bonacchi, S.; Genovese, D.; Juris, R.; Montalti, M.; Prodi, L.; Rampazzo, E.; Zaccheroni, N. Luminescent silica nanoparticles: Extending the frontiers of brightness. Angew. Chem. Int. Ed. 2011, 50, 4056–4066. [Google Scholar] [CrossRef]
- Yamada, K.; Nagashima, I.; Hachisu, M.; Matsuo, I.; Shimizu, H. Efficient solid-phase synthesis of cyclic RGD peptides under controlled microwave heating. Tetrahedron Lett. 2012, 53, 1066–1070. [Google Scholar] [CrossRef]
- Greco, A.; Tani, S.; De Marco, R.; Gentilucci, L. Synthesis and analysis of the conformational preferences of 5-aminomethyloxazolidine-2,4-dione scaffolds: First examples of beta2-and beta2,2-homo-Freidinger lactam analogues. Chem. Eur. J. 2014, 20, 13390–13404. [Google Scholar] [CrossRef]
- Malesevic, M.; Strijowski, U.; Bächle, D.; Sewald, N. An improved method for the solution cyclization of peptides under pseudo-high dilution conditions. J. Biotechnol. 2004, 112, 73–77. [Google Scholar] [CrossRef]
- Rampazzo, E.; Bonacchi, S.; Juris, R.; Genovese, D.; Prodi, L.; Zaccheroni, N.; Montalti, M. Dual-mode, anisotropy-encoded, ratiometric fluorescent nanosensors: Towards multiplexed detection. Chem. Eur. J. 2018, 24, 16743–16746. [Google Scholar] [CrossRef]
- Rampazzo, E.; Bonacchi, S.; Juris, R.; Montalti, M.; Genovese, D.; Zaccheroni, N.; Prodi, L.; Rambaldi, D.C.; Zattoni Andrea, C.; Reschiglian, P. Energy transfer from silica core–surfactant shell nanoparticles to hosted molecular fluorophores. J. Phys. Chem. B 2010, 114, 14605–14613. [Google Scholar] [CrossRef]
- Valenti, G.; Rampazzo, E.; Bonacchi, S.; Khajvand, T.; Juris, R.; Montalti, M.; Marcaccio, M.; Paolucci, F.; Prodi, L. A versatile strategy for tuning the color of electrochemiluminescence using silica nanoparticles. Chem. Commun. 2012, 48, 4187–4189. [Google Scholar] [CrossRef]
- Haubner, R.; Finsinger, D.; Kessler, H. Stereoisomeric peptide libraries and peptidomimetics for designing selective inhibitors of the αvβ3 integrin for a new cancer therapy. Angew. Chem. Int. Ed. 1997, 36, 1374–1389. [Google Scholar] [CrossRef]
- Adamou, R.; Coly, A.; Douabalé, S.E.; Saleck, M.L.; Gaye-Seye, M.D.; Tine, A. Fluorimetric determination of histamine in fish using micellar media and fluorescamine as labelling reagent. J. Fluoresc. 2005, 15, 679–688. [Google Scholar] [CrossRef]
- Parolin, C.; Frisco, G.; Foschi, C.; Giordani, B.; Salvo, M.; Vitali, B.; Marangoni, A.; Calonghi, N. Lactobacillus crispatus BC5 interferes with chlamydia trachomatis infectivity through integrin modulation in cervical cells. Front. Microbiol. 2018, 9, 2630. [Google Scholar] [CrossRef]
- Kemperman, H.; Wijnands, Y.M.; Roos, E. αV Integrins on HT-29 colon carcinoma cells: Adhesion to fibronectin is mediated solely by small amounts of αVβ6, and αVβ5 is codistributed with actin fibers. Exp. Cell Res. 1997, 234, 156–164. [Google Scholar] [CrossRef]
- Schmidt, R.; Streit, M.; Kaiser, R.; Herzberg, F.; Schirner, M.; Schramm, K.; Kaufmann, C.; Henneken, M.; Schäfer-Korting, M.; Thiel, E.; et al. De novo expression of the α5β1-fibronectin receptor in HT29 colon-cancer cells reduces activity of c-src. increase of c-src activity by attachment on fibronectin. Int. J. Cancer 1998, 76, 91–98. [Google Scholar] [CrossRef]
- Storm, G.; Belliot, S.O.; Daemen, T.; Lasic, D.D. Surface modification of nanoparticles to oppose uptake by the mononuclear phagocyte system. Adv. Drug Deliv. Rev. 1995, 17, 31–48. [Google Scholar] [CrossRef]
- Huo, Q.; Liu, J.; Wang, L.Q.; Jiang, Y.; Lambert, T.N.; Fang, E. A new class of silica cross-linked micellar core-shell nanoparticles. J. Am. Chem. Soc. 2006, 128, 6447–6453. [Google Scholar] [CrossRef]
- Rampazzo, E.; Boschi, F.; Bonacchi, S.; Juris, R.; Montalti, M.; Zaccheroni, N.; Prodi, L.; Calderan, L.; Rossi, B.; Becchi, S.; et al. Multicolor core/shell silica nanoparticles for in vivo and ex vivo imaging. Nanoscale 2012, 4, 824–830. [Google Scholar] [CrossRef]
- Helle, M.; Rampazzo, E.; Monchanin, M.; Marchal, F.; Guillemin, F.; Bonacchi, S.; Salis, F.; Prodi, L.; Bezdetnaya, L. Surface chemistry architecture of silica nanoparticles determine the efficiency of in vivo fluorescence lymph node mapping. ACS Nano 2013, 7, 8645–8657. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Marco, R.; Rampazzo, E.; Zhao, J.; Prodi, L.; Paolillo, M.; Picchetti, P.; Gallo, F.; Calonghi, N.; Gentilucci, L. Integrin-Targeting Dye-Doped PEG-Shell/Silica-Core Nanoparticles Mimicking the Proapoptotic Smac/DIABLO Protein. Nanomaterials 2020, 10, 1211. https://0-doi-org.brum.beds.ac.uk/10.3390/nano10061211
De Marco R, Rampazzo E, Zhao J, Prodi L, Paolillo M, Picchetti P, Gallo F, Calonghi N, Gentilucci L. Integrin-Targeting Dye-Doped PEG-Shell/Silica-Core Nanoparticles Mimicking the Proapoptotic Smac/DIABLO Protein. Nanomaterials. 2020; 10(6):1211. https://0-doi-org.brum.beds.ac.uk/10.3390/nano10061211
Chicago/Turabian StyleDe Marco, Rossella, Enrico Rampazzo, Junwei Zhao, Luca Prodi, Mayra Paolillo, Pierre Picchetti, Francesca Gallo, Natalia Calonghi, and Luca Gentilucci. 2020. "Integrin-Targeting Dye-Doped PEG-Shell/Silica-Core Nanoparticles Mimicking the Proapoptotic Smac/DIABLO Protein" Nanomaterials 10, no. 6: 1211. https://0-doi-org.brum.beds.ac.uk/10.3390/nano10061211