Enhancing Antibodies’ Binding Capacity through Oriented Functionalization of Plasmonic Surfaces
Abstract
:1. Introduction

2. Materials and Methods
2.1. Fabrication of Plasmonic Nanoparticles Clusters
2.2. Scanning Electron Microscopy (SEM) Images of Samples
2.3. Atomic Force Microscopy (AFM) Images of Samples
2.4. Functionalization of Gold Nanoparticles Clusters
2.5. Raman Spectroscopy Analysis of Samples
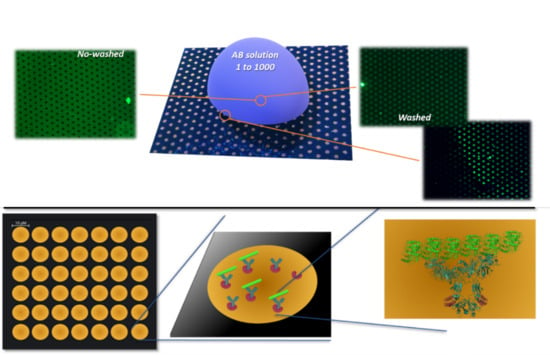
2.6. Acquiring Fluorescence Images of Samples
2.7. Fluorescence Testing on Antibody Immobilization
3. Results
3.1. Clusters of Plasmonic Gold Nanoparticles
3.2. Atomic Force Microscopy
3.3. Fluorescence Analysis for MEF Effect Evaluation
3.4. Characterization of the Functionalized Plasmonic Gold Nanoparticles
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Leavy, O. Therapeutic antibodies: Past, present and future. Nat. Rev. Immunol. 2010, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Steinitz, M. Three decades of human monoclonal antibodies: Past, present and future developments. Hum. Antibodies 2009, 18, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Research Antibodies Market Size, Share & Trends Analysis Report by Type (Monoclonal, Polyclonal), by End Use, by Product, by Technology, by Source, by Application (Oncology, Stem Cells), and Segment Forecasts, 2020–2027; Research and Markets: Dublin, Ireland, 2020; p. 142.
- Shen, M.; Rusling, J.F.; Dixit, C.K. Site-selective orientated immobilization of antibodies and conjugates for immunodiagnostics development. Methods 2017, 116, 95–111. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.; Lichtman, A.; Pillai, S. Cellular and Molecular Immunology, 9th ed.; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Choe, W.; Durgannavar, T.A.; Chung, S.J. Fc-Binding Ligands of Immunoglobulin G: An Overview of High Affinity Proteins and Peptides. Materials 2016, 9, 994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, M.; Shankar, V.; Vafai, A. Therapeutic Applications of Monoclonal Antibodies. Am. J. Med. Sci. 2002, 324, 14–30. [Google Scholar] [CrossRef]
- Mullard, A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Discov. 2021. [Google Scholar] [CrossRef]
- Dixit, C.K.; Vashist, S.K.; O’Neill, F.T.; O’Reilly, B.; MacCraith, B.D.; O’Kennedy, R. Development of a High Sensitivity Rapid Sandwich ELISA Procedure and Its Comparison with the Conventional Approach. Anal. Chem. 2010, 82, 7049–7052. [Google Scholar] [CrossRef]
- Dixit, C.K.; Vashist, S.K.; MacCraith, B.D.; O’Kennedy, R. Multisubstrate-compatible ELISA procedures for rapid and high-sensitivity immunoassays. Nat. Protoc. 2011, 6, 439–445. [Google Scholar] [CrossRef]
- Correa, I.; Ilieva, K.M.; Crescioli, S.; Lombardi, S.; Figini, M.; Cheung, A.; Spicer, J.; Tutt, A.N.J.; Nestle, F.O.; Karagiannis, P.; et al. Evaluation of Antigen-Conjugated Fluorescent Beads to Identify Antigen-Specific B Cells. Front. Immunol. 2018, 9, 493. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Lin, Q.; Huang, L.; Zhai, Y.; Liu, Y.; Deng, Y.; Su, E.; He, N. Ultrasensitive chemiluminescence immunoassay with enhanced precision for the detection of cTnI amplified by acridinium ester-loaded microspheres and internally calibrated by magnetic fluorescent nanoparticles. Nanoscale 2021, 13, 3275–3284. [Google Scholar] [CrossRef]
- Vancini, R.; Wang, G.; Ferreira, D.; Hernandez, R.; Brown, D.T. Alphavirus Genome Delivery Occurs Directly at the Plasma Membrane in a Time- and Temperature-Dependent Process. J. Virol. 2013, 87, 4352–4359. [Google Scholar] [CrossRef] [Green Version]
- Neri, L.M.; Santi, S.; Cinti, C.; Sabatelli, P.; Valmori, A.; Capanni, C.; Capitani, S.; Stuppia, L.; Maraldi, N.M. Multiple fluorescence and reflectance simultaneous detection by confocal microscopy of HaeIII digested DNA sequences. Eur. J. Cell Biol. 1996, 71, 120–128. [Google Scholar] [PubMed]
- Wang, D.; Dai, Y.; Wang, X.; Yu, P.; Qu, S.; Liu, Z.; Cao, Y.; Zhang, L.; Ping, Y.; Liu, W.; et al. Determination of plasma β-amyloids by rolling circle amplification chemiluminescent immunoassay for noninvasive diagnosis of Alzheimer’s disease. Microchim. Acta 2021, 188, 1–9. [Google Scholar] [CrossRef]
- Liang, L.-H.; Cheng, X.; Yu, H.-L.; Yang, Y.; Mu, X.-H.; Chen, B.; Li, X.-S.; Wu, J.-N.; Yan, L.; Liu, C.-C.; et al. Quantitative detection of ricin in beverages using trypsin/Glu-C tandem digestion coupled with ultra-high-pressure liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2021, 413, 585–597. [Google Scholar] [CrossRef]
- Khattar, N.; Yablonska, S.; Baranov, S.V.; Baranova, O.V.; Kretz, E.S.; Larkin, T.M.; Carlisle, D.L.; Richardson, R.; Friedlander, R.M. Isolation of functionally active and highly purified neuronal mitochondria from human cortex. J. Neurosci. Methods 2016, 263, 1–6. [Google Scholar] [CrossRef]
- Cetin, D.; Okan, M.; Bat, E.; Kulah, H. A comparative study on EpCAM antibody immobilization on gold surfaces and microfluidic channels for the detection of circulating tumor cells. Colloids Surfaces B Biointerfaces 2020, 188, 110808. [Google Scholar] [CrossRef] [PubMed]
- Gallo, V.; Lai, A.; Pasquo, A.; Almaviva, S.; Iacobelli, S.; Persichetti, L.; Capellini, G.; Antonini, G. Surface-enhanced Raman scattering (SERS)–based immunosystem for ultrasensitive detection of the 90K biomarker. Anal. Bioanal. Chem. 2020, 412, 7659–7667. [Google Scholar] [CrossRef]
- Laschi, S.; Mascini, M.; Scortichini, G.; Fránek, M.; Mascini, M. Polychlorinated Biphenyls (PCBs) Detection in Food Samples Using an Electrochemical Immunosensor. J. Agric. Food Chem. 2003, 51, 1816–1822. [Google Scholar] [CrossRef]
- Dontha, N.; Nowall, W.B.; Kuhr, W.G. Development of sub-micron patterned carbon electrodes for immunoassays. J. Pharm. Biomed. Anal. 1999, 19, 83–91. [Google Scholar] [CrossRef]
- Harmsen, M.M.; Fijten, H.P.D. Improved Functional Immobilization of Llama Single-domain Antibody Fragments to Polystyrene Surfaces Using Small Peptides. J. Immunoass. Immunochem. 2012, 33, 234–251. [Google Scholar] [CrossRef]
- Cho, I.-H.; Paek, E.-H.; Lee, H.; Kang, J.Y.; Kim, T.S.; Paek, S.-H. Site-directed biotinylation of antibodies for controlled immobilization on solid surfaces. Anal. Biochem. 2007, 365, 14–23. [Google Scholar] [CrossRef]
- Franco, E.J.; Hofstetter, H.; Hofstetter, O. A comparative evaluation of random and site-specific immobilization techniques for the preparation of antibody-based chiral stationary phases. J. Sep. Sci. 2006, 29, 1458–1469. [Google Scholar] [CrossRef]
- Trilling, A.K.; Harmsen, M.M.; Ruigrok, V.J.; Zuilhof, H.; Beekwilder, J. The effect of uniform capture molecule orientation on biosensor sensitivity: Dependence on analyte properties. Biosens. Bioelectron. 2013, 40, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Baio, J.E.; Cheng, F.; Ratner, D.M.; Stayton, P.S.; Castner, D.G. Probing orientation of immobilized humanized anti-lysozyme variable fragment by time-of-flight secondary-ion mass spectrometry. J. Biomed. Mater. Res. Part A 2011, 97A, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makaraviciute, A.; Ramanaviciene, A. Site-directed antibody immobilization techniques for immunosensors. Biosens. Bioelectron. 2013, 50, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Billah, M.; Hays, H.C.W.; Millner, P.A. Development of a myoglobin impedimetric immunosensor based on mixed self-assembled monolayer onto gold. Microchim. Acta 2008, 160, 447–454. [Google Scholar] [CrossRef]
- Mattos, A.; Freitas, T.; Silva, V.; Dutra, R. A dual quartz crystal microbalance for human cardiac troponin T in real time detection. Sensors Actuators B Chem. 2012, 161, 439–446. [Google Scholar] [CrossRef]
- Pulido-Tofiño, P.; Barrero-Moreno, J.; Pérez-Conde, M. Flow-through fluoroimmunosensor for isoproturon determination in agricultural foodstuff: Evaluation of antibody immobilization on solid support. Anal. Chim. Acta 2000, 417, 85–94. [Google Scholar] [CrossRef]
- Tran, Q.H.; Nguyen, T.H.H.; Mai, A.T.; Vu, Q.K.; Phan, T.N.; Nguyen, T.T. Development of electrochemical immunosensors based on different serum antibody immobilization methods for detection of Japanese encephalitis virus. Adv. Nat. Sci. Nanosci. Nanotechnol. 2012, 3, 015012. [Google Scholar] [CrossRef] [Green Version]
- Buijs, J.; Norde, W.; Lichtenbelt, J.W.T. Changes in the Secondary Structure of Adsorbed IgG and F(ab‘)2 Studied by FTIR Spectroscopy. Langmuir 1996, 12, 1605–1613. [Google Scholar] [CrossRef]
- Buijs, J.; Lichtenbelt, J.W.; Norde, W.; Lyklema, J. Adsorption of monoclonal IgGs and their F(ab′)2 fragments onto polymeric surfaces. Colloids Surfaces B Biointerfaces 1995, 5, 11–23. [Google Scholar] [CrossRef]
- Sun, X.; Yu, G.; Xu, Q.; Li, N.; Xiao, C.; Yin, X.; Cao, K.; Han, J.; He, Q.-Y. Putative cobalt- and nickel-binding proteins and motifs in Streptococcus pneumoniae. Metallomics 2013, 5, 928–935. [Google Scholar] [CrossRef]
- Haugland, R.P.; You, W.W.; Robert, M.J. Coupling of Antibodies with Biotin. Avidin-Biot. Interact. 2008, 418, 13–24. [Google Scholar] [CrossRef]
- Buchwalow, I.B.; Samoilova, V.; Boecker, W.; Tiemann, M. Non-specific binding of antibodies in immunohistochemistry: Fallacies and facts. Sci. Rep. 2011, 1, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.-A.A.; Hsu, W.-L.; Liao, W.-C.; Chiu, J.-K.; Chen, M.-L.; Chang, H.-C.; Li, C.-C. Ultrasensitive electrochemical detection of biotin using electrically addressable site-oriented antibody immobilization approach via aminophenyl boronic acid. Biosens. Bioelectron. 2010, 26, 1021–1027. [Google Scholar] [CrossRef]
- O’Shannessy, D.J.; Quarles, R.H. Specific conjugation reactions of the oligosaccharide moieties of immunoglobulins. J. Appl. Biochem. 1985, 7, 347–355. [Google Scholar] [PubMed]
- Wolfe, C.; Hage, D. Studies on the Rate and Control of Antibody Oxidation by Periodate. Anal. Biochem. 1995, 231, 123–130. [Google Scholar] [CrossRef]
- Hudson, B.G.; Barker, R. The overoxidation of carbohydrates with sodium metaperiodate. J. Org. Chem. 1967, 32, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- Peluso, P.; Wilson, D.S.; Do, D.; Tran, H.; Venkatasubbaiah, M.; Quincy, D.; Heidecker, B.; Poindexter, K.; Tolani, N.; Phelan, M.; et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003, 312, 113–124. [Google Scholar] [CrossRef]
- Tsai, W.-C.; Pai, P.-J.R. Surface plasmon resonance-based immunosensor with oriented immobilized antibody fragments on a mixed self-assembled monolayer for the determination of staphylococcal enterotoxin B. Microchim. Acta 2009, 166, 115–122. [Google Scholar] [CrossRef]
- Rouet, R.; Lowe, D.; Dudgeon, K.; Roome, B.; Schofield, P.; Langley, D.; Andrews, J.; Whitfeld, P.; Jermutus, L.; Christ, D. Expression of high-affinity human antibody fragments in bacteria. Nat. Protoc. 2012, 7, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Kausaite-Minkstimiene, A.; Ramanaviciene, A.; Kirlyte, J.; Ramanavicius, A. Comparative Study of Random and Oriented Antibody Immobilization Techniques on the Binding Capacity of Immunosensor. Anal. Chem. 2010, 82, 6401–6408. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Muguruma, H.; Karube, I. A Novel Method of Immobilizing Antibodies on a Quartz Crystal Microbalance Using Plasma-Polymerized Films for Immunosensors. Anal. Chem. 1996, 68, 1695–1700. [Google Scholar] [CrossRef] [PubMed]
- Vikholm-Lundin, I. Immunosensing Based on Site-Directed Immobilization of Antibody Fragments and Polymers that Reduce Nonspecific Binding. Langmuir 2005, 21, 6473–6477. [Google Scholar] [CrossRef]
- DeSilva, B.S.; Wilson, G.S. Solid phase synthesis of bifunctional antibodies. J. Immunol. Methods 1995, 188, 9–19. [Google Scholar] [CrossRef]
- Lu, B.; Xie, J.; Lu, C.; Wu, C.; Wei, Y. Oriented Immobilization of Fab’ Fragments on Silica Surfaces. Anal. Chem. 1995, 67, 83–87. [Google Scholar] [CrossRef]
- Bonroy, K.; Frederix, F.; Reekmans, G.; Dewolf, E.; De Palma, R.; Borghs, G.; Declerck, P.; Goddeeris, B. Comparison of random and oriented immobilisation of antibody fragments on mixed self-assembled monolayers. J. Immunol. Methods 2006, 312, 167–181. [Google Scholar] [CrossRef]
- Quintero-Hernández, V.; Juárez-González, V.R.; Ortíz-León, M.; Sánchez, R.; Possani, L.D.; Becerril, B. The change of the scFv into the Fab format improves the stability and in vivo toxin neutralization capacity of recombinant antibodies. Mol. Immunol. 2007, 44, 1307–1315. [Google Scholar] [CrossRef]
- O’Brien, J.C.; Jones, V.W.; Porter, M.D.; Mosher, C.L.; Henderson, E. Immunosensing Platforms Using Spontaneously Adsorbed Antibody Fragments on Gold. Anal. Chem. 2000, 72, 703–710. [Google Scholar] [CrossRef] [Green Version]
- Vikholm, I. Self-assembly of antibody fragments and polymers onto gold for immunosensing. Sensors Actuators B Chem. 2005, 106, 311–316. [Google Scholar] [CrossRef]
- Song, H.Y.; Zhou, X.; Hobley, J.; Su, X. Comparative Study of Random and Oriented Antibody Immobilization as Measured by Dual Polarization Interferometry and Surface Plasmon Resonance Spectroscopy. Langmuir 2012, 28, 997–1004. [Google Scholar] [CrossRef]
- Deisenhofer, J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-.ANG. resolution. Biochemistry 1981, 20, 2361–2370. [Google Scholar] [CrossRef]
- Sauer-Eriksson, A.E.; Kleywegt, G.J.; Uhlén, M.; Jones, T.A. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 1995, 3, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Quinn, J.; Patel, P.; Fitzpatrick, B.; Manning, B.; Dillon, P.; Daly, S.; O’Kennedy, R.; Alcocer, M.; Lee, H.; Morgan, M.; et al. The use of regenerable, affinity ligand-based surfaces for immunosensor applications. Biosens. Bioelectron. 1999, 14, 587–595. [Google Scholar] [CrossRef]
- Goding, J.W. Use of staphylococcal protein A as an immunological reagent. J. Immunol. Methods 1978, 20, 241–253. [Google Scholar] [CrossRef]
- Majzik, A.; Patakfalvi, R.; Hornok, V.; Dékány, I. Growing and stability of gold nanoparticles and their functionalization by cysteine. Gold Bull. 2009, 42, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Candeloro, P.; Grande, E.; Raimondo, R.; Di Mascolo, D.; Gentile, F.; Coluccio, M.L.; Perozziello, G.; Malara, N.; Francardi, M.; Di Fabrizio, E. Raman database of amino acids solutions: A critical study of Extended Multiplicative Signal Correction. Analyst 2013, 138, 7331–7340. [Google Scholar] [CrossRef] [PubMed]
- Gentile, F.; Tirinato, L.; Battista, E.; Causa, F.; Liberale, C.; di Fabrizio, E.M.; Decuzzi, P. Cells preferentially grow on rough substrates. Biomaterials 2010, 31, 7205–7212. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.E.J.; McCourt, M.R. SERS enhancement by aggregated Au colloids: Effect of particle size. Phys. Chem. Chem. Phys. 2009, 11, 7455–7462. [Google Scholar] [CrossRef]
- Karunakaran, C.; Bhargava, K.; Benjamin, R. Biosensors and Bioelectronics; Elsevier: Amsterdam, The Netherlands, 2018; pp. 102–116. [Google Scholar]
- Tian, F.; Bonnier, F.; Casey, A.; Shanahan, A.E.; Byrne, H. Surface enhanced Raman scattering with gold nanoparticles: Effect of particle shape. Anal. Methods 2014, 6, 9116–9123. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Yi, J.; Hu, S.; Liu, B.-J.; Liu, J.; Wang, X.; Ren, B. Size Effect on SERS of Gold Nanorods Demonstrated via Single Nanoparticle Spectroscopy. J. Phys. Chem. C 2016, 120, 20806–20813. [Google Scholar] [CrossRef]
- Ventura, B.D.; Gelzo, M.; Battista, E.; Alabastri, A.; Schirato, A.; Castaldo, G.; Corso, G.; Gentile, F.; Velotta, R. Biosensor for Point-of-Care Analysis of Immunoglobulins in Urine by Metal Enhanced Fluorescence from Gold Nanoparticles. ACS Appl. Mater. Interfaces 2019, 11, 3753–3762. [Google Scholar] [CrossRef] [PubMed]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Ettah, I.; Ashton, L. Engaging with Raman Spectroscopy to Investigate Antibody Aggregation. Antibodies 2018, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Fagnano, C.; Fini, G. Antibody—Antigen interactions studied by means of Raman Spectroscopy. J. Mol. Struct. 1993, 294, 111–114. [Google Scholar] [CrossRef]
- Ahmadivand, A.; Gerislioglu, B.; Tomitaka, A.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Extreme sensitive metasensor for targeted biomarkers identification using colloidal nanoparticles-integrated plasmonic unit cells. Biomed. Opt. Express 2018, 9, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Xie, L.; Zhu, J.; Xu, X.; Ye, Z.; Wang, C.; Ma, Y.; Ying, Y. Gold Nanoparticle-Based Terahertz Metamaterial Sensors: Mechanisms and Applications. ACS Photon 2016, 3, 2308–2314. [Google Scholar] [CrossRef]
- Hermanson, G.T. Bioconjugate Techniques; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Limongi, T.; Schipani, R.; Di Vito, A.; Giugni, A.; Francardi, M.; Torre, B.; Allione, M.; Miele, E.; Malara, N.; Alrasheed, S.; et al. Photolithography and micromolding techniques for the realization of 3D polycaprolactone scaffolds for tissue engineering applications. Microelectron. Eng. 2015, 141, 135–139. [Google Scholar] [CrossRef]
- Romano, G.; Mancini, R.; Fedele, P.; Curigliano, G.; Flamini, G.; Giovagnoli, M.R.; Malara, N.; Boninsegna, A.; Vecchione, A.; Santella, R.M.; et al. Immunohistochemical analysis of 4-aminobiphenyl-DNA adducts in oral mucosal cells of smokers and nonsmokers. Anticancer. Res. 1997, 17, 2827–2830. [Google Scholar]
- Malara, N.; Leotta, A.; Sidoti, A.; Lio, S.; D’Angelo, R.; Caparello, B.; Munao, F.; Pino, F.; Amato, A. Ageing, hormonal behaviour and cyclin D1 in ductal breast carcinomas. Breast 2006, 15, 81–89. [Google Scholar] [CrossRef]
- Perozziello, G.; Catalano, R.; Francardi, M.; Rondanina, E.; Pardeo, F.; De Angelis, F.; Malara, N.; Candeloro, P.; Morrone, G.; Di Fabrizio, E. A microfluidic device integrating plasmonic nanodevices for Raman spectroscopy analysis on trapped single living cells. Microelectron. Eng. 2013, 111, 314–319. [Google Scholar] [CrossRef]
- Onesto, V.; Cancedda, L.; Coluccio, M.L.; Nanni, M.; Pesce, M.; Malara, N.; Cesarelli, M.; Di Fabrizio, E.; Amato, F.; Gentile, F. Nano-topography Enhances Communication in Neural Cells Networks. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Aquila, I.; Sacco, M.A.; Abenavoli, L.; Malara, N.; Arena, V.; Grassi, S.; Ausania, F.; Boccuto, L.; Ricci, C.; Gratteri, S.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Pandemic. Arch. Pathol. Lab. Med. 2020, 144, 1048–1056. [Google Scholar] [CrossRef]
- Perozziello, G.; Simone, G.; Candeloro, P.; Gentile, F.; Malara, N.; Larocca, R.; Coluccio, M.L.; Pullano, S.A.; Tirinato, L.; Geschke, O.; et al. A Fluidic Motherboard for Multiplexed Simultaneous and Modular Detection in Microfluidic Systems for Biological Application. Micro Nanosyst. 2010, 2, 227–238. [Google Scholar] [CrossRef] [Green Version]







| Immobilization Format | Methodology | Advantages | Disadvantages |
|---|---|---|---|
| Random | Covalent attachment via amine coupling Physiosorption | In some cases shows good sensitivity [28,29] Surface regeneration for multiple analyses after covalent attachment [30] | Lower sensitivity in comparison to site-directed immobilization methods [31] In case of physiosorption denaturation of proteins, very low stability and random protein orientation [32,33] |
| Via His-tag | Expression of recombinant antibody with His-tag |
|
|
| Via biotinylated antibody | Site specific biotinylation |
|
|
| Via an oxidized oligosaccharide moiety | Chemical or enzymatic oxidation of an oligosaccharide moiety and coupling to amine or hydrazine terminated supports |
| |
| Via antibody fragments | Chemical reduction or genetic engineering based disruption of disulfide bridges and immobilization via sulfhydryl groups | ||
| Site-directed Via Fc binding proteins | Affinity interactions with a preformed layer of proteins specific to the Fc regions of Ab, e.g., proteins A, G, A/G, L, anti-Fc, recombinant proteins |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coluccio, M.L.; Grillo, F.; Onesto, V.; Garo, V.; Scala, C.; Cuzzola, P.; Calfa, M.; Candeloro, P.; Gentile, F.; Piletsky, S.; et al. Enhancing Antibodies’ Binding Capacity through Oriented Functionalization of Plasmonic Surfaces. Nanomaterials 2021, 11, 2620. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11102620
Coluccio ML, Grillo F, Onesto V, Garo V, Scala C, Cuzzola P, Calfa M, Candeloro P, Gentile F, Piletsky S, et al. Enhancing Antibodies’ Binding Capacity through Oriented Functionalization of Plasmonic Surfaces. Nanomaterials. 2021; 11(10):2620. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11102620
Chicago/Turabian StyleColuccio, Maria Laura, Fabiana Grillo, Valentina Onesto, Virginia Garo, Cinzia Scala, Paola Cuzzola, Michela Calfa, Patrizio Candeloro, Francesco Gentile, Sergey Piletsky, and et al. 2021. "Enhancing Antibodies’ Binding Capacity through Oriented Functionalization of Plasmonic Surfaces" Nanomaterials 11, no. 10: 2620. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11102620