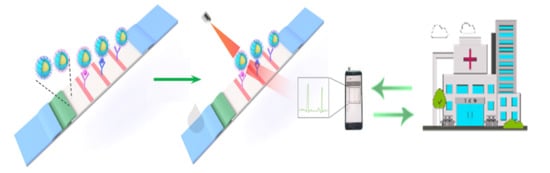
Simultaneous Detection of Inflammatory Biomarkers by SERS Nanotag-Based Lateral Flow Assay with Portable Cloud Raman Spectrometer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
2.3. Synthesis of Core-Shell AuMBA@mSiO2 Nanoparticles
2.4. Preparation of Core-Shell AuMBA@mSiO2 SERS Nanotags
2.5. Preparation of SERS-LFA Strip
2.6. Optimization of the SERS-LFA Strip
2.7. Procedures of Simultaneous and Quantitative Detection of SAA and CRP by SERS-LFA Strip
3. Results and Discussion
3.1. Fabrication and Characterization of Core-Shell AuMBA@mSiO2 SERS Nanotags
3.2. Raman Signal of AuMBA@mSiO2 SERS Nanotags
3.3. Sensitivity and Specificity of the SERS-LFA Strip
3.4. Simultaneous and Quantitative Detection of CRP and SAA Based on the SERS-LFA Strip
3.5. Simultaneous and Quantitative Detection of CRP and SAA by Portable Cloud Raman Spectrometer for POCT
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kankala, R.K.; Zhang, H.; Liu, C.-G.; Kanubaddi, K.R.; Lee, C.-H.; Wang, S.-B.; Cui, W.; Santos, H.A.; Lin, K.; Chen, A.-Z. Metal species–encapsulated mesoporous silica nanoparticles: Current advancements and latest breakthroughs. Adv. Funct. Mater. 2019, 29, 1902652. [Google Scholar] [CrossRef]
- Serebrennikova, K.V.; Samsonova, J.V.; Osipov, A.P. A semi-quantitative rapid multi-range gradient lateral flow immunoassay for procalcitonin. Microchim. Acta 2019, 186, 423. [Google Scholar] [CrossRef]
- Kim, S.-W.; Cho, I.-H.; Lim, G.-S.; Park, G.-N.; Paek, S.-H. Biochemical-immunological hybrid biosensor based on two-dimensional chromatography for on-site sepsis diagnosis. Biosens. Bioelectron. 2017, 98, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, A.L.; Vanimaya; Ravindran, S.; Saikant, R.; Lakshmi, S.; Kartik, R.; Manoj, G. Procalcitonin: A promising diagnostic marker for sepsis and antibiotic therapy. J. Intensive Care 2017, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Larsen, F.F.; Petersen, J.A. Novel biomarkers for sepsis: A narrative review. Eur. J. Intern. Med. 2017, 45, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, X.; Li, K.; Liu, H.; Xiao, R.; Wang, W.; Wang, C.; Wang, S. Fe3O4@Au SERS tags-based lateral flow assay for simultaneous detection of serum amyloid A and C-reactive protein in unprocessed blood sample. Sens. Actuators B Chem. 2020, 320, 128350. [Google Scholar] [CrossRef]
- Talamona, F.; Truffi, M.; Caldarone, A.A.; Ricciardi, A.; Corsi, F.; Pellegrini, G.; Morasso, C.; Taglietti, A. Stable and scalable SERS tags conjugated with neutravidin for the detection of fibroblast activation protein (FAP) in primary fibroblasts. Nanotechnology 2021, 32, 295703. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, X.; Gu, B.; Liu, H.; Xiao, R.; Wang, C.; Wang, S. Quantitative and simultaneous detection of two inflammation biomarkers via a fluorescent lateral flow immunoassay using dual-color SiO2@QD nanotags. Microchim. Acta 2020, 187, 1–11. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, H.; Yang, Y.; Yan, R.; Zhao, Y.; Wang, Y.; Chen, A.; Shao, S.; Jiang, P.; Li, Y.-Q. Bacterial species-identifiable magnetic nanosystems for early sepsis diagnosis and extracorporeal photodynamic blood disinfection. Nanoscale 2018, 10, 132–141. [Google Scholar] [CrossRef]
- Yi, J.; Qin, Q.; Wang, Y.; Zhang, R.; Bi, H.; Yu, S.; Liu, B.; Qiao, L. Identification of pathogenic bacteria in human blood using IgG-modified Fe3O4 magnetic beads as a sorbent and MALDI-TOF MS for profiling. Microchim. Acta 2018, 185, 1–10. [Google Scholar] [CrossRef]
- Xianyu, Y.; Wu, J.; Chen, Y.; Zheng, W.; Xie, M.; Jiang, X. Controllable Assembly of Enzymes for Multiplexed Lab-on-a-Chip Bioassays with a Tunable Detection Range. Angew. Chem. Int. Ed. 2018, 57, 7503–7507. [Google Scholar] [CrossRef] [PubMed]
- Belushkin, A.; Yesilkoy, F.; Gonzalez-Lopez, J.J.; Ruiz-Rodriguez, J.C.; Ferrer, R.; Fabrega, A.; Altug, H. Rapid and Digital Detection of Inflammatory Biomarkers Enabled by a Novel Portable Nanoplasmonic Imager. Small 2020, 16, 1906108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.-H.; Ahn, J.-H.; Kim, J.-Y.; Choi, J.-M.; Lim, K.-C.; Park, T.J.; Heo, N.S.; Lee, H.G.; Kim, J.-W.; Choi, Y.-K. CRP detection from serum for chip-based point-of-care testing system. Biosens. Bioelectron. 2013, 41, 322–327. [Google Scholar] [CrossRef]
- van Houten, C.; Groot, J.A.H.D.; Klein, A.; Srugo, I.; Chistyakov, I.; De Waal, W.; Meijssen, C.B.; Avis, W.; Wolfs, T.F.W.; Shachor-Meyouhas, Y.; et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): A double-blind, multicentre, validation study. Lancet Infect. Dis. 2017, 17, 431–440. [Google Scholar] [CrossRef]
- Kim, H.; Lee, J.U.; Song, S.; Kim, S.; Sim, S.J. A shape-code nanoplasmonic biosensor for multiplex detection of Alzheimer’s disease biomarkers. Biosens. Bioelectron. 2018, 101, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, H.-Z.; Zhu, X.; Su, J.-Q.; Ren, B.; Zhu, Y.-G.; Cui, L. Rapid antibiotic susceptibility testing of pathogenic bacteria using heavy-water-labeled single-cell raman spectroscopy in clinical samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef]
- Shen, H.; Wang, J.; Liu, H.; Li, Z.; Jiang, F.-L.; Wang, F.-B.; Yuan, Q. Rapid and selective detection of pathogenic bacteria in bloodstream infections with aptamer-based recognition. ACS Appl. Mater. Interfaces 2016, 8, 19371–19378. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Ni, H.; Sun, L.; Su, E.; Chen, H.; Gu, Z.; Zhao, X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018, 106, 204–211. [Google Scholar] [CrossRef]
- Jia, X.; Wang, C.; Rong, Z.; Li, J.; Wang, K.; Qie, Z.; Xiao, R.; Wang, S. Dual dye-loaded Au@Ag coupled to a lateral flow immunoassay for the accurate and sensitive detection of Mycoplasma pneumoniae infection. RSC Adv. 2018, 8, 21243–21251. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Huang, L.; Liu, B.; Su, E.; Chen, H.-Y.; Gu, Z.; Zhao, X. Quantitative detection of multiplex cardiac biomarkers with encoded SERS nanotags on a single T line in lateral flow assay. Sens. Actuators B Chem. 2018, 277, 502–509. [Google Scholar] [CrossRef]
- Lin, B.; Guan, Z.; Song, Y.; Song, E.; Lu, Z.; Liu, D.; An, Y.; Zhu, Z.; Zhou, L.; Yang, C.J. Lateral flow assay with pressure meter readout for rapid point-of-care detection of disease-associated protein. Lab Chip 2018, 18, 965–970. [Google Scholar] [CrossRef]
- Huang, L.; Tian, S.; Zhao, W.; Liu, K.; Ma, X.; Guo, J. Multiplexed detection of biomarkers in lateral-flow immunoassays. Analyst 2020, 145, 2828–2840. [Google Scholar] [CrossRef]
- Blanco-Covián, L.; Montes-García, V.; Girard, A.; Fernández-Abedul, M.; Pérez-Juste, J.; Pastoriza-Santos, I.; Faulds, K.; Graham, D.; Blanco-López, M.C. Au@Ag SERRS tags coupled to a lateral flow immunoassay for the sensitive detection of pneumolysin. Nanoscale 2017, 9, 2051–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for Sensitive and Simultaneous Detection of Respiratory Viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tian, S.; Liu, K.; Guo, J. IoT-enabled fluorescence sensor for quantitative KET detection and anti-drug situational awareness. IEEE Trans. Nanobiosci. 2021, 20, 2–8. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, R.; Han, L.; Tu, B.; Zhao, D. One-pot synthesis of thermally stable gold@mesoporous silica core-shell nanospheres with catalytic activity. Nano Res. 2013, 6, 871–879. [Google Scholar] [CrossRef]
- Liu, H.-B.; Chen, C.-Y.; Zhang, C.-N.; Du, X.-J.; Li, P.; Wang, S. Functionalized Au-MBA@Ag nanoparticles as an optical and SERS dual probe in a lateral flow strip for the quantitative detection of escherichia coli O157:H7. J. Food Sci. 2019, 84, 2916–2924. [Google Scholar] [CrossRef]
- Deng, D.; Yang, H.; Liu, C.; Zhao, K.; Li, J.; Deng, A. Ultrasensitive detection of diclofenac in water samples by a novel surface-enhanced Raman scattering (SERS)-based immunochromatographic assay using AgMBA@SiO2-Ab as immunoprobe. Sens. Actuators B Chem. 2019, 283, 563–570. [Google Scholar] [CrossRef]
- Song, S.; Liu, N.; Zhao, Z.; Ediage, E.N.; Wu, S.; Sun, C.; De Saeger, S.; Wu, A. Multiplex lateral flow immunoassay for mycotoxin determination. Anal. Chem. 2014, 86, 4995–5001. [Google Scholar] [CrossRef]
- Gul, O.; Calay, E.; Sezerman, U.; Basaga, H.; Gurbuz, Y. Sandwich-type, antibody microarrays for the detection and quantification of cardiovascular risk markers. Sens. Actuators B Chem. 2007, 125, 581–588. [Google Scholar] [CrossRef]
- Rong, Z.; Xiao, R.; Xing, S.; Xiong, G.; Yu, Z.; Wang, L.; Jia, X.; Wang, K.; Cong, Y.; Wang, S. SERS-based lateral flow assay for quantitative detection of C-reactive protein as an early bio-indicator of a radiation-induced inflammatory response in nonhuman primates. Analyst 2018, 143, 2115–2121. [Google Scholar] [CrossRef]
- Qi, X.; Huang, Y.; Lin, Z.; Xu, L.; Yunye, H. Dual-quantum-dots-labeled lateral flow strip rapidly quantifies procalcitonin and C-reactive protein. Nanoscale Res. Lett. 2016, 11, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.; Wang, F.; Li, N.; Wu, R.; Li, J.; Shen, H.; Li, L.S.; Guo, F. Development of dual quantum dots-based fluorescence-linked immunosorbent assay for simultaneous detection on inflammation biomarkers. Sens. Actuators B Chem. 2019, 301, 127118. [Google Scholar] [CrossRef]





| Detection method | Analyte | LOD | Time | Reference |
|---|---|---|---|---|
| Protein microarrays | SAA | 5.9 ng/mL | >2.5 h | Gul et al., 2007 [31] |
| SERS-LFA strip | CRP | 0.01 ng/mL | 20 min | Rong et al., 2018 [32] |
| Magnetoimmunosensor | CRP | 8 ng/mL | 20 min | Fernández et al., 2016 [33] |
| Fluorescent-LFA strip | CRP | 0.5 ng/mL | 15 min | Yang et al., 2020 [8] |
| SERS-LFA strip | SAA, CRP | 0.1, 0.01 ng/mL | 30 min | Liu et al., 2020 [6] |
| QD-based FLISA | SAA, CRP | 2.39, 6.37 ng/mL | >1 h | Lv et al., 2019 [34] |
| SERS-LFA strip | SAA, CRP | 0.1, 0.05 ng/mL | 20 min | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Liu, X.; Guo, J.; Zhang, Y.; Guo, J.; Wu, X.; Wang, B.; Ma, X. Simultaneous Detection of Inflammatory Biomarkers by SERS Nanotag-Based Lateral Flow Assay with Portable Cloud Raman Spectrometer. Nanomaterials 2021, 11, 1496. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11061496
Li Y, Liu X, Guo J, Zhang Y, Guo J, Wu X, Wang B, Ma X. Simultaneous Detection of Inflammatory Biomarkers by SERS Nanotag-Based Lateral Flow Assay with Portable Cloud Raman Spectrometer. Nanomaterials. 2021; 11(6):1496. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11061496
Chicago/Turabian StyleLi, Yang, Xiaojia Liu, Jiuchuan Guo, Yueting Zhang, Jinhong Guo, Xinggui Wu, Bo Wang, and Xing Ma. 2021. "Simultaneous Detection of Inflammatory Biomarkers by SERS Nanotag-Based Lateral Flow Assay with Portable Cloud Raman Spectrometer" Nanomaterials 11, no. 6: 1496. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11061496