DNAzyme-Amplified Electrochemical Biosensor Coupled with pH Meter for Ca2+ Determination at Variable pH Environments
Abstract
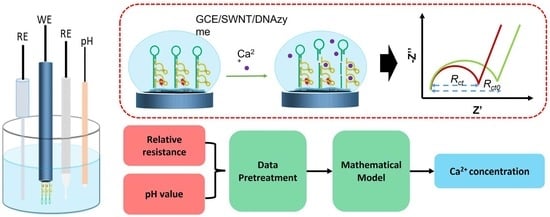
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Apparatus and Measurements
2.3. Fabrication of the Biosensor
2.4. Electrochemical Measurements
2.5. Sensing of Real Samples
3. Results
3.1. Choice of Materials
3.2. Characterization
3.3. CV and EIS Characterization
3.4. Optimization
3.5. Selectivity
3.6. Ca2+ Sensing
3.7. Mathematical Model
3.7.1. Stepwise Linear Regression
3.7.2. Neural Network Fitting
3.7.3. Gaussian Process Regression
3.8. Comparison of Detection Performance
3.9. Detection of Ca2+ in Real Sample
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Balk, E.M.; for the International Osteoporosis Foundation Calcium Steering Committee; Adam, G.P.; Langberg, V.N.; Earley, A.; Clark, P.; Ebeling, P.R.; Mithal, A.; Rizzoli, R.; Zerbini, C.A.F.; et al. Global dietary calcium intake among adults: A systematic review. Osteoporos. Int. 2017, 28, 3315–3324. [Google Scholar] [CrossRef] [Green Version]
- Giorgi, C.; Marchi, S.; Pinton, P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 2018, 19, 713–730. [Google Scholar] [CrossRef]
- Maltsev, V.; Lakatta, E.G. Dynamic interactions of an intracellular Ca2+ clock and membrane ion channel clock underlie robust initiation and regulation of cardiac pacemaker function. Cardiovasc. Res. 2008, 77, 274–284. [Google Scholar] [CrossRef]
- Xu, Y.; Ye, J.; Zhou, D.; Su, L. Research progress on applications of calcium derived from marine organisms. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Daresjö, S. Determinants for Milk Fever: An Epidemiological Study of Swedish Dairy Cows. 2020. Available online: https://stud.epsilon.slu.se/15725/1/daresjo_s_200120.pdf (accessed on 13 December 2021).
- Tesfaye, A. Calcium requirement in relation to milk fever for high yielding dairy cows: A review. J. Food Agric. Environ. 2019, 18, 1–12. [Google Scholar]
- Kaufman, E.; Asselstine, V.; LeBlanc, S.; Duffield, T.; Devries, T. Association of rumination time and health status with milk yield and composition in early-lactation dairy cows. J. Dairy Sci. 2018, 101, 462–471. [Google Scholar] [CrossRef]
- Yue, J.; Li, L.; Cao, L.; Zan, M.; Yang, D.; Wang, Z.; Chang, Z.; Mei, Q.; Miao, P.; Dong, W.-F. Two-step hydrothermal preparation of carbon dots for calcium ion detection. ACS Appl. Mater. Interfaces 2019, 11, 44566–44572. [Google Scholar] [CrossRef]
- Hernández-Castellano, L.E.; Hernandez, L.L.; Weaver, S.; Bruckmaier, R.M. Increased serum serotonin improves parturient calcium homeostasis in dairy cows. J. Dairy Sci. 2017, 100, 1580–1587. [Google Scholar] [CrossRef]
- Goff, J.P.; Hohman, A.; Timms, L.L. Effect of subclinical and clinical hypocalcemia and dietary cation-anion difference on rumination activity in periparturient dairy cows. J. Dairy Sci. 2020, 103, 2591–2601. [Google Scholar] [CrossRef] [PubMed]
- Nedić, S.; Palamarević, M.; Arsić, S.; Jovanović, L.; Prodanović, R.; Kirovski, D.; Vujanac, I. Parathyroid hormone response in treatment of subclinical hypocalcemia in postpartum dairy cows. Res. Vet. Sci. 2020, 132, 351–356. [Google Scholar] [CrossRef]
- Ibrahim, N.; Kirmani, M.A. Milk Fever in Dairy Cows: A Systematic Review. Available online: https://www.researchgate.net/profile/Nuraddis-Ibrahim/publication/350942379_ (accessed on 13 December 2021).
- Uniyal, S.; Sharma, R.K. Technological advancement in electrochemical biosensor based detection of organophosphate pesticide chlorpyrifos in the environment: A review of status and prospects. Biosens. Bioelectron. 2018, 116, 37–50. [Google Scholar] [CrossRef]
- Beitollahi, H.; Mohammadi, S.Z.; Safaei, M.; Tajik, S. Applications of electrochemical sensors and biosensors based on modified screen-printed electrodes: A review. Anal. Methods 2020, 12, 1547–1560. [Google Scholar] [CrossRef]
- Liang, G.; Man, Y.; Li, A.; Jin, X.; Liu, X.; Pan, L. DNAzyme-based biosensor for detection of lead ion: A review. Microchem. J. 2017, 131, 145–153. [Google Scholar] [CrossRef]
- Zhang, J. RNA-cleaving DNAzymes: Old catalysts with new tricks for intracellular and in vivo applications. Catalysts 2018, 8, 550. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Saran, R.; Ding, J.; Liu, J. Two completely different mechanisms for highly specific Na+ recognition by DNAzymes. ChemBioChem 2017, 18, 1828–1835. [Google Scholar] [CrossRef]
- He, Y.; Chen, D.; Huang, P.-J.J.; Zhou, Y.; Ma, L.; Xu, K.; Yang, R.; Liu, J. Misfolding of a DNAzyme for ultrahigh sodium selectivity over potassium. Nucleic Acids Res. 2018, 46, 10262–10271. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, Y.; Liu, G. Label-free biosensor using a silver specific RNA-cleaving DNAzyme functionalized single-walled carbon nanotube for silver ion determination. Nanomaterials 2018, 8, 258. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, Y.; Liu, G. Electrochemical biosensor using DNA embedded phosphorothioate modified RNA for mercury ion determination. ACS Sens. 2018, 3, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.-S.; Li, B.-C.; Zhou, Y.-L.; Wang, H.-Y.; Wang, M.-H.; Ai, S.-Y. Signal-on fluorescence biosensor for microRNA-21 detection based on DNA strand displacement reaction and Mg2+-dependent DNAzyme cleavage. Biosens. Bioelectron. 2017, 96, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Wang, J.; Xiong, B.; Hou, X. Electrochemical impedance biosensor array based on DNAzyme-functionalized single-walled carbon nanotubes using Gaussian process regression for Cu (II) and Hg (II) determination. Microchim. Acta 2020, 187, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Huang, P.J.; He, M.; Lyu, M.; Wang, C.; Wang, S.; Liu, J. Sensitivity of a classic DNAzyme for Pb2+ modulated by cations, anions and buffers. Analyst 2020, 145, 1384–1388. [Google Scholar] [CrossRef]
- Zhu, P.; Shang, Y.; Tian, W.; Huang, K.; Luo, Y.; Xu, W. Ultra-sensitive and absolute quantitative detection of Cu2+ based on DNAzyme and digital PCR in water and drink samples. Food Chem. 2017, 221, 1770–1777. [Google Scholar] [CrossRef]
- Gong, L.; Zhao, Z.; Lv, Y.-F.; Huan, S.-Y.; Fu, T.; Zhang, X.-B.; Shen, G.-L.; Yu, R.-Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. 2015, 51, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Huang, P.-J.J.; de Rochambeau, D.; Moon, W.J.; Zhang, J.; Lyu, M.; Wang, S.; Sleiman, H.; Liu, J. Selection of a metal ligand modified DNAzyme for detecting Ni2+. Biosens. Bioelectron. 2020, 165, 112285. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, Q.; Zhao, Y.; Nan, X.; Zhang, F.; Wang, Y.; Wang, Y.; Hua, D.; Zheng, S.; Jiang, L.; et al. Electrochemical device based on nonspecific DNAzyme for the high-accuracy determination of Ca2+ with Pb2+ interference. Bioelectrochemistry 2021, 140, 107732. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Saran, R.; Huang, P.-J.J.; Ding, J.; Liu, J. An exceptionally selective DNA cooperatively binding two Ca2+ ions. ChemBioChem 2017, 18, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhou, W.; Liu, J. Screening of DNAzyme mutants for highly sensitive and selective detection of calcium in milk. Anal. Methods 2018, 10, 1740–1746. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, S.; Cong, X.; Sun, S.; Wu, J.-B.; Zhang, D.; Yang, F.; Yang, J.; Tan, P.-H.; Li, Y. Electronic Raman Scattering in Suspended Semiconducting Carbon Nanotube. J. Phys. Chem. Lett. 2020, 11, 10497–10503. [Google Scholar] [CrossRef]
- Dugasani, S.R.; Gnapareddy, B.; Kesama, M.R.; Ha, T.H.; Park, S.H. DNA and DNA–CTMA composite thin films embedded with carboxyl group-modified multi-walled carbon nanotubes. J. Ind. Eng. Chem. 2018, 68, 79–86. [Google Scholar] [CrossRef]
- Zhou, N.; Pierre, J.W.; Trudnowski, D. A stepwise regression method for estimating dominant electromechanical modes. IEEE Trans. Power Syst. 2011, 27, 1051–1059. [Google Scholar] [CrossRef]
- Wang, H.; Ramnani, P.; Pham, T.; Villarreal, C.C.; Yu, X.; Liu, G.; Mulchandani, A. Gas biosensor arrays based on single-stranded DNA-functionalized single-walled carbon nanotubes for the detection of volatile organic compound biomarkers released by huanglongbing disease-infected citrus trees. Sensors 2019, 19, 4795. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Jin, H.; Chen, X.; Dai, J.; Wang, L.; Zhang, D. Soft sensor development for online quality prediction of industrial batch rubber mixing process using ensemble just-in-time Gaussian process regression models. Chemom. Intell. Lab. 2016, 155, 170–182. [Google Scholar] [CrossRef]
- Deringer, V.L.; Bartók, A.P.; Bernstein, N.; Wilkins, D.M.; Ceriotti, M.; Csányi, G. Gaussian Process Regression for Materials and Molecules. Chem. Rev. 2021, 121, 10073–10141. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Zhou, J.; Xu, J.; Fan, M.; Ma, Y.; Zhou, M.; Li, T.; Zhao, S. A smartphone-based colorimetry after dispersive liquid–liquid microextraction for rapid quantification of calcium in water and food samples. Microchem. J. 2019, 149, 104072. [Google Scholar] [CrossRef]
- Shibata, H.; Ikeda, Y.; Hiruta, Y.; Citterio, D. Inkjet-printed pH-independent paper-based calcium sensor with fluorescence signal readout relying on a solvatochromic dye. Anal. Bioanal. Chem. 2020, 412, 3489–3497. [Google Scholar] [CrossRef]
- Liu, S.; Ding, J.; Qin, W. Current pulse based ion-selective electrodes for chronopotentiometric determination of calcium in seawater. Anal. Chim. Acta 2018, 1031, 67–74. [Google Scholar] [CrossRef]
- Ocaña, C.; Abramova, N.; Bratov, A.; Lindfors, T.; Bobacka, J. Calcium-selective electrodes based on photo-cured polyurethane-acrylate membranes covalently attached to methacrylate functionalized poly (3, 4-ethylenedioxythiophene) as solid-contact. Talanta 2018, 186, 279–285. [Google Scholar] [CrossRef]
- Ahmad, R.; Tripathy, N.; Ahn, M.-S.; Yoo, J.-Y.; Hahn, Y.-B. Preparation of a highly conductive seed layer for calcium sensor fabrication with enhanced sensing performance. ACS Sens. 2018, 3, 772–778. [Google Scholar] [CrossRef]













| DNA Name | Sequences and Modifications (Starting from 5 Terminal ′) | |
|---|---|---|
| Ca_sub | NO-substrate | NH2-(CH26)GCGGTAGAAGGATATCACTGAGCACTG |
| NS-substrate | NH2-(C6)GCGGTAGAAGG/rA/TATCACTGAGCACTG | |
| DS-substrate | NH2-(C6)GCGGTAGAAGG/rA/TATCACTGAGCACTGGG/rA/TAAGCGG TAGAACTCACAATGTATAATGCGCGCATTATACATTGTGAGT | |
| CAZyme | EtNa-C5T | CAGTGCTCAGTGATTGTTGGAATGGCTCATGCCACACTCTTTTCTACCGC |
| sEtNa-C5T | TCTACCGCTTATCCCAGTGCTCAGTGATTGTTGGAATGGCTC ATGCCACACTCTTTTCTACCGC | |
| dEtNa-C5T | TCTACCGCTTTGTTGGAATGGCTCATGCCACACTCTTCAGTGCT CAGTGATTGTTGGAATGGCTCATGCCACACTCTTTTCTACCGC | |
| Sensor | Method | pH | Time (min) | Linear Range (μM) | LOD (μM) | Ref. |
|---|---|---|---|---|---|---|
| CD-EGTA | Fluorescent | 7 | 240 | 15 ∼ 300 μM | 0.38 μM | [8] |
| DNAzyme/SWNT/FET | Current | 6.9 | 9 | 10 ∼ 1000 | 7.2 | [27] |
| DLLME | UV-visible | 12.8 | 30 | 1.5 ∼ 37.5 | 0.425 | [36] |
| SD-ISO | Fluorescence | 6.0–8.0 | 5 | 10 ∼ 106 | 9.3 | [37] |
| Ca-ISE | Potential | - | 1.67 | 10−2.5 ∼ 10−1.5 M | - | [38] |
| Ca2+-SCISE | Potential | - | 1 | 10−1 ∼ 104 | 5 | [39] |
| Fe2O3-ZnO NRs/FET | Current | 7.6 | - | 0.01 ∼ 3.0 mM | 0.05 | [40] |
| Electrochemical device | EIS | 4.0–7.5 | 7 | 5 ∼ 2.5 × 104 | 4.2 | This work |
| Sample | Electrochemical Device (mM) | AAS (mM) | Recovery (%) | |
|---|---|---|---|---|
| Blood | 1 | 0.78 | 0.73 | 106.85 |
| 2 | 1.15 | 1.12 | 102.68 | |
| 3 | 0.81 | 0.88 | 92.05 | |
| 4 | 1.09 | 1.17 | 93.16 | |
| Milk | 1 | 27.32 | 29.20 | 93.56 |
| 2 | 31.34 | 28.45 | 105.34 | |
| 3 | 21.87 | 22.78 | 96.01 | |
| 4 | 34.34 | 37.87 | 90.68 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhang, F.; Wang, Y.; Shi, F.; Luo, Q.; Zheng, S.; Chen, J.; Dai, D.; Yang, L.; Tang, X.; et al. DNAzyme-Amplified Electrochemical Biosensor Coupled with pH Meter for Ca2+ Determination at Variable pH Environments. Nanomaterials 2022, 12, 4. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12010004
Wang H, Zhang F, Wang Y, Shi F, Luo Q, Zheng S, Chen J, Dai D, Yang L, Tang X, et al. DNAzyme-Amplified Electrochemical Biosensor Coupled with pH Meter for Ca2+ Determination at Variable pH Environments. Nanomaterials. 2022; 12(1):4. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12010004
Chicago/Turabian StyleWang, Hui, Fan Zhang, Yue Wang, Fangquan Shi, Qingyao Luo, Shanshan Zheng, Junhong Chen, Dingzhen Dai, Liang Yang, Xiangfang Tang, and et al. 2022. "DNAzyme-Amplified Electrochemical Biosensor Coupled with pH Meter for Ca2+ Determination at Variable pH Environments" Nanomaterials 12, no. 1: 4. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12010004