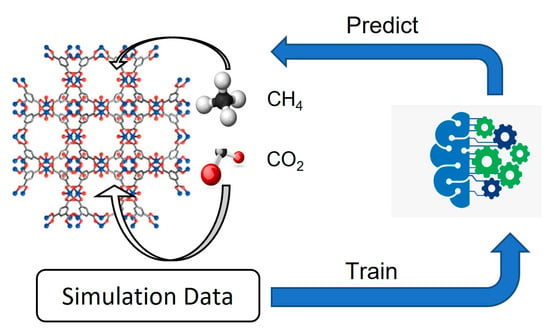
Deep Learning Models for Predicting Gas Adsorption Capacity of Nanomaterials
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Preparation
2.2. Deep Learning Algorithms
2.3. Model Development
2.4. Model Evaluation
3. Results and Discussion
3.1. Prediction of Methane Adsorption of MOFs
3.2. Prediction of Carbon Dioxide Adsorption of MOFs
3.3. Model Transferability
3.4. Models Constructed Using a Mixture of MOFs and COFs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Begum, S.; Karim, A.N.M.; Ansari, M.N.M.; Hashmi, M.S.J. Nanomaterials. In Encyclopedia of Renewable and Sustainable Materials; Hashmi, S., Choudhury, I.A., Eds.; Elsevier: Oxford, UK, 2020; pp. 515–539. [Google Scholar]
- Hofmann-Amtenbrink, M.; Grainger, D.W.; Hofmann, H. Nanoparticles in medicine: Current challenges facing inorganic nanoparticle toxicity assessments and standardizations. Nanomedicine 2015, 11, 1689–1694. [Google Scholar] [CrossRef] [PubMed]
- Chellaram, C.; Murugaboopathi, G.; John, A.A.; Sivakumar, R.; Ganesan, S.; Krithika, S.; Priya, G. Significance of Nanotechnology in Food Industry. APCBEE Procedia 2014, 8, 109–113. [Google Scholar] [CrossRef]
- Pomerantseva, E.; Bonaccorso, F.; Feng, X.; Cui, Y.; Gogotsi, Y. Energy Storage: The Future Enabled by Nanomaterials. Science 2019, 366, eaan8285. [Google Scholar] [CrossRef]
- Yu, H.; Li, L.; Zhang, Y. Silver Nanoparticle-based Thermal Interface Materials with Ultra-low Thermal Resistance for Power Electronics Applications. Scr. Mater. 2012, 66, 931–934. [Google Scholar] [CrossRef]
- Chen, X.; Gianneschi, N.; Ginger, D.; Nam, J.-M.; Zhang, H. Programmable Materials. Adv. Mater. 2021, 33, 2107344. [Google Scholar] [CrossRef] [PubMed]
- Batten, S.R.; Champness, N.R.; Chen, X.-M.; Garcia-Martinez, J.; Kitagawa, S.; Öhrström, L.; O’Keeffe, M.; Paik Suh, M.; Reedijk, J. Terminology of metal–organic frameworks and coordination polymers (IUPAC Recommendations 2013). Pure Appl. Chem. 2013, 85, 1715–1724. [Google Scholar] [CrossRef]
- Gomollón-Bel, F. Ten Chemical Innovations That Will Change Our World: IUPAC identifies emerging technologies in Chemistry with potential to make our planet more sustainable. Chem. Int. 2019, 41, 12–17. [Google Scholar] [CrossRef]
- Ahmed, A.; Seth, S.; Purewal, J.; Wong-Foy, A.G.; Veenstra, M.; Matzger, A.J.; Siegel, D.J. Exceptional hydrogen storage achieved by screening nearly half a million metal-organic frameworks. Nat. Commun. 2019, 10, 1568. [Google Scholar] [CrossRef] [PubMed]
- Alezi, D.; Belmabkhout, Y.; Suyetin, M.; Bhatt, P.M.; Weseliński, Ł.J.; Solovyeva, V.; Adil, K.; Spanopoulos, I.; Trikalitis, P.N.; Emwas, A.-H.; et al. MOF Crystal Chemistry Paving the Way to Gas Storage Needs: Aluminum-Based soc-MOF for CH4, O2, and CO2 Storage. J. Am. Chem. Soc. 2015, 137, 13308–13318. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Zuluaga, S.; Gong, Q.; Gao, Y.; Nijem, N.; Li, J.; Thonhauser, T.; Chabal, Y.J. Competitive Coadsorption of CO2 with H2O, NH3, SO2, NO, NO2, N2, O2, and CH4 in M-MOF-74 (M = Mg, Co, Ni): The Role of Hydrogen Bonding. Chem. Mater. 2015, 27, 2203–2217. [Google Scholar] [CrossRef] [Green Version]
- Chong, S.; Lee, S.; Kim, B.; Kim, J. Applications of machine learning in metal-organic frameworks. Coord. Chem. Rev. 2020, 423, 213487. [Google Scholar] [CrossRef]
- Anderson, R.; Rodgers, J.; Argueta, E.; Biong, A.; Gómez-Gualdrón, D.A. Role of Pore Chemistry and Topology in the CO2 Capture Capabilities of MOFs: From Molecular Simulation to Machine Learning. Chem. Mater. 2018, 30, 6325–6337. [Google Scholar] [CrossRef]
- Evans, J.D.; Fraux, G.; Gaillac, R.; Kohen, D.; Trousselet, F.; Vanson, J.-M.; Coudert, F.-X. Computational Chemistry Methods for Nanoporous Materials. Chem. Mater. 2017, 29, 199–212. [Google Scholar] [CrossRef]
- Altintas, C.; Altundal, O.F.; Keskin, S.; Yildirim, R. Machine Learning Meets with Metal Organic Frameworks for Gas Storage and Separation. J. Chem. Inf. Model. 2021, 61, 2131–2146. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tong, W.; Fang, H.; Xie, Q.; Hong, H.; Perkins, R.; Wu, J.; Tu, M.; Blair, R.M.; Branham, W.S.; et al. An Integrated "4-phase" Approach for Setting Endocrine Disruption Screening Priorities-Phase I and II Predictions of Estrogen Receptor Binding Affinity. SAR QSAR Environ. Res. 2002, 13, 69–88. [Google Scholar] [CrossRef]
- Cheng, F.; Hong, H.; Yang, S.; Wei, Y. Individualized network-based drug repositioning infrastructure for precision oncology in the panomics era. Brief. Bioinform. 2017, 18, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xu, L.; Fang, H.; Richard, A.M.; Bray, J.D.; Judson, R.S.; Zhou, G.; Colatsky, T.J.; Aungst, J.L.; Teng, C.; et al. EADB: An Estrogenic Activity Database for Assessing Potential Endocrine Activity. Toxicol. Sci. 2013, 135, 277–291. [Google Scholar] [CrossRef]
- Hong, H.; Neamati, N.; Winslow, H.E.; Christensen, J.L.; Orr, A.; Pommier, Y.; Milne, G.W. Identification of HIV-1 integrase inhibitors based on a four-point pharmacophore. Antivir. Chem. Chemother. 1998, 9, 461–472. [Google Scholar] [CrossRef]
- Ng, H.W.; Zhang, W.; Shu, M.; Luo, H.; Ge, W.; Perkins, R.; Tong, W.; Hong, H. Competitive Molecular Docking Approach for Predicting Estrogen Receptor Subtype α Agonists and Antagonists. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef]
- Selvaraj, C.; Sakkiah, S.; Tong, W.; Hong, H. Molecular Dynamics Simulations and Applications in Computational Toxicology and Nanotoxicology. Food Chem. Toxicol. 2018, 112, 495–506. [Google Scholar] [CrossRef]
- Sakkiah, S.; Guo, W.; Pan, B.; Ji, Z.; Yavas, G.; Azevedo, M.; Hawes, J.; Patterson, T.A.; Hong, H. Elucidating Interactions Between SARS-CoV-2 Trimeric Spike Protein and ACE2 Using Homology Modeling and Molecular Dynamics Simulations. Front. Chem. 2021, 8, 622632. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Wang, X.; Hong, H.; Benfenati, E.; Giesy, J.P.; Gini, G.C.; Kusko, R.; Zhang, X.; Yu, H.; Shi, W. Structures of Endocrine-Disrupting Chemicals Determine Binding to and Activation of the Estrogen Receptor α and Androgen Receptor. Environ. Sci. Technol. 2020, 54, 11424–11433. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Zhu, J.; Chen, M.; Gong, P.; Zhang, C.; Tong, W. Quantitative Structure–Activity Relationship Models for Predicting Risk of Drug-Induced Liver Injury in Humans. In Drug-Induced Liver Toxicity; Chen, M., Will, Y., Eds.; Springer: New York, NY, USA, 2018; pp. 77–100. [Google Scholar]
- Luo, H.; Ye, H.; Ng, H.W.; Shi, L.; Tong, W.; Mendrick, D.L.; Hong, H. Machine Learning Methods for Predicting HLA-Peptide Binding Activity. Bioinform. Biol. Insights 2015, 9, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Thakkar, S.; Chen, M.; Tong, W. Development of Decision Forest Models for Prediction of Drug-Induced Liver Injury in Humans Using A Large Set of FDA-approved Drugs. Sci. Rep. 2017, 7, 17311. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.W.; Doughty, S.W.; Luo, H.; Ye, H.; Ge, W.; Tong, W.; Hong, H. Development and Validation of Decision Forest Model for Estrogen Receptor Binding Prediction of Chemicals Using Large Data Sets. Chem. Res. Toxicol. 2015, 28, 2343–2351. [Google Scholar] [CrossRef]
- Sakkiah, S.; Selvaraj, C.; Gong, P.; Zhang, C.; Tong, W.; Hong, H. Development of Estrogen Receptor beta Binding Prediction Model Using Large Sets of Chemicals. Oncotarget 2017, 8, 92989–93000. [Google Scholar] [CrossRef]
- Huang, Y.; Li, X.; Xu, S.; Zheng, H.; Zhang, L.; Chen, J.; Hong, H.; Kusko, R.; Li, R. Quantitative Structure–Activity Relationship Models for Predicting Inflammatory Potential of Metal Oxide Nanoparticles. Environ. Health Perspect. 2020, 128, 067010. [Google Scholar] [CrossRef]
- Hong, H.; Tong, W.; Xie, Q.; Fang, H.; Perkins, R. An in silico ensemble method for lead discovery: Decision forest. SAR QSAR Environ. Res. 2005, 16, 339–347. [Google Scholar] [CrossRef]
- Ng, H.W.; Shu, M.; Luo, H.; Ye, H.; Ge, W.; Perkins, R.; Tong, W.; Hong, H. Estrogenic activity data extraction and in silico prediction show the endocrine disruption potential of bisphenol A replacement compounds. Chem. Res. Toxicol. 2015, 28, 1784–1795. [Google Scholar] [CrossRef]
- Jablonka, K.M.; Ongari, D.; Moosavi, S.M.; Smit, B. Big-Data Science in Porous Materials: Materials Genomics and Machine Learning. Chem. Rev. 2020, 120, 8066–8129. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, L.; Li, S.; Liang, H.; Qiao, Z. Adsorption Behavior of Metal-Organic Frameworks: From Single Simulation, High-throughput Computational Screening to Machine Learning. Comput. Mater. Sci. 2021, 193, 110383. [Google Scholar] [CrossRef]
- Deng, X.; Yang, W.; Li, S.; Liang, H.; Shi, Z.; Qiao, Z. Large-Scale Screening and Machine Learning to Predict the Computation-Ready, Experimental Metal-Organic Frameworks for CO2 Capture from Air. Appl. Sci. 2020, 10, 569. [Google Scholar] [CrossRef]
- Fanourgakis, G.S.; Gkagkas, K.; Tylianakis, E.; Klontzas, E.; Froudakis, G. A Robust Machine Learning Algorithm for the Prediction of Methane Adsorption in Nanoporous Materials. J. Phys. Chem. A 2019, 123, 6080–6087. [Google Scholar] [CrossRef] [PubMed]
- Fanourgakis, G.S.; Gkagkas, K.; Tylianakis, E.; Froudakis, G.E. A Universal Machine Learning Algorithm for Large-Scale Screening of Materials. J. Am. Chem. Soc. 2020, 142, 3814–3822. [Google Scholar] [CrossRef]
- Lee, S.; Kim, B.; Cho, H.; Lee, H.; Lee, S.Y.; Cho, E.S.; Kim, J. Computational Screening of Trillions of Metal–Organic Frameworks for High-Performance Methane Storage. ACS Appl. Mater. Interfaces 2021, 13, 23647–23654. [Google Scholar] [CrossRef]
- Anderson, R.; Biong, A.; Gómez-Gualdrón, D.A. Adsorption Isotherm Predictions for Multiple Molecules in MOFs Using the Same Deep Learning Model. J. Chem. Theory Comput. 2020, 16, 1271–1283. [Google Scholar] [CrossRef]
- Ma, R.; Colón, Y.J.; Luo, T. Transfer Learning Study of Gas Adsorption in Metal–Organic Frameworks. ACS Appl. Mater. Interfaces 2020, 12, 34041–34048. [Google Scholar] [CrossRef]
- Wang, R.; Zhong, Y.; Bi, L.; Yang, M.; Xu, D. Accelerating Discovery of Metal–Organic Frameworks for Methane Adsorption with Hierarchical Screening and Deep Learning. ACS Appl. Mater. Interfaces 2020, 12, 52797–52807. [Google Scholar] [CrossRef]
- Wilmer, C.E.; Leaf, M.; Lee, C.Y.; Farha, O.K.; Hauser, B.G.; Hupp, J.T.; Snurr, R.Q. Large-scale Screening of Hypothetical Metal–Organic Frameworks. Nat. Chem. 2012, 4, 83–89. [Google Scholar] [CrossRef]
- Pardakhti, M.; Moharreri, E.; Wanik, D.; Suib, S.L.; Srivastava, R. Machine Learning Using Combined Structural and Chemical Descriptors for Prediction of Methane Adsorption Performance of Metal Organic Frameworks (MOFs). ACS Comb. Sci. 2017, 19, 640–645. [Google Scholar] [CrossRef]
- Wilmer, C.E.; Farha, O.K.; Bae, Y.-S.; Hupp, J.T.; Snurr, R.Q. Structure–property relationships of porous materials for carbon dioxide separation and capture. Energy Environ. Sci. 2012, 5, 9849–9856. [Google Scholar] [CrossRef]
- Mercado, R.; Fu, R.-S.; Yakutovich, A.V.; Talirz, L.; Haranczyk, M.; Smit, B. In Silico Design of 2D and 3D Covalent Organic Frameworks for Methane Storage Applications. Chem. Mater. 2018, 30, 5069–5086. [Google Scholar] [CrossRef]
- Fernandez, M.; Woo, T.K.; Wilmer, C.E.; Snurr, R.Q. Large-Scale Quantitative Structure–Property Relationship (QSPR) Analysis of Methane Storage in Metal–Organic Frameworks. J. Phys. Chem. C 2013, 117, 7681–7689. [Google Scholar] [CrossRef]
- Willems, T.F.; Rycroft, C.H.; Kazi, M.; Meza, J.C.; Haranczyk, M. Algorithms and Tools for High-throughput Geometry-based Analysis of Crystalline Porous Materials. Microporous Mesoporous Mater. 2012, 149, 134–141. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, M.; Zhu, X.; Zhao, L. Attention-based LSTM for Aspect-level Sentiment Classification. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 606–615. [Google Scholar]
- Rao, G.; Huang, W.; Feng, Z.; Cong, Q. LSTM with sentence representations for document-level sentiment classification. Neurocomputing 2018, 308, 49–57. [Google Scholar] [CrossRef]
- Fernandez, M.; Trefiak, N.R.; Woo, T.K. Atomic Property Weighted Radial Distribution Functions Descriptors of Metal–Organic Frameworks for the Prediction of Gas Uptake Capacity. J. Phys. Chem. C 2013, 117, 14095–14105. [Google Scholar] [CrossRef]
- Hu, K.; Mischo, H. Modeling High-Pressure Methane Adsorption on Shales with a Simplified Local Density Model. ACS Omega 2020, 5, 5048–5060. [Google Scholar] [CrossRef]
- Ursueguía, D.; Díaz, E.; Ordóñez, S. Adsorption of methane and nitrogen on Basolite MOFs: Equilibrium and kinetic studies. Microporous Mesoporous Mater. 2020, 298, 110048. [Google Scholar] [CrossRef]
- He, Y.; Zhou, W.; Qian, G.; Chen, B. Methane storage in metal–organic frameworks. Chem. Soc. Rev. 2014, 43, 5657–5678. [Google Scholar] [CrossRef]
- Peng, Y.; Krungleviciute, V.; Eryazici, I.; Hupp, J.T.; Farha, O.K.; Yildirim, T. Methane Storage in Metal–Organic Frameworks: Current Records, Surprise Findings, and Challenges. J. Am. Chem. Soc. 2013, 135, 11887–11894. [Google Scholar] [CrossRef] [Green Version]
- Bucior, B.J.; Bobbitt, N.S.; Islamoglu, T.; Goswami, S.; Gopalan, A.; Yildirim, T.; Farha, O.K.; Bagheri, N.; Snurr, R.Q. Energy-based descriptors to rapidly predict hydrogen storage in metal–organic frameworks. Mol. Syst. Des. Eng. 2019, 4, 162–174. [Google Scholar] [CrossRef]
- Horcajada, P.; Gref, R.; Baati, T.; Allan, P.K.; Maurin, G.; Couvreur, P.; Férey, G.; Morris, R.E.; Serre, C. Metal–Organic Frameworks in Biomedicine. Chem. Rev. 2012, 112, 1232–1268. [Google Scholar] [CrossRef] [PubMed]
- Dureckova, H.; Krykunov, M.; Aghaji, M.Z.; Woo, T.K. Robust Machine Learning Models for Predicting High CO2 Working Capacity and CO2 /H2 Selectivity of Gas Adsorption in Metal Organic Frameworks for Precombustion Carbon Capture. J. Phys. Chem. C 2019, 123, 4133–4139. [Google Scholar] [CrossRef]






| Gas | Pressure (bar) | MOFs | Mean Adsorption | Standard Deviation |
|---|---|---|---|---|
| CO2 | 0.05 | 70,433 | 2.2466 | 5.3255 |
| CO2 | 0.5 | 70,433 | 37.365 | 35.3544 |
| CO2 | 2.5 | 70,433 | 92.9512 | 56.5359 |
| CH4 | 1 | 70,608 | 17.8184 | 17.7097 |
| CH4 | 5.8 | 28,417 | 57.5549 | 33.442 |
| CH4 | 35 | 70,605 | 139.1942 | 44.8701 |
| CH4 | 65 | 27,151 | 172.11 | 50.1996 |
| CH4 | 65 | 17,098 * | 153.3413 | 37.7033 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Liu, J.; Dong, F.; Chen, R.; Das, J.; Ge, W.; Xu, X.; Hong, H. Deep Learning Models for Predicting Gas Adsorption Capacity of Nanomaterials. Nanomaterials 2022, 12, 3376. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12193376
Guo W, Liu J, Dong F, Chen R, Das J, Ge W, Xu X, Hong H. Deep Learning Models for Predicting Gas Adsorption Capacity of Nanomaterials. Nanomaterials. 2022; 12(19):3376. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12193376
Chicago/Turabian StyleGuo, Wenjing, Jie Liu, Fan Dong, Ru Chen, Jayanti Das, Weigong Ge, Xiaoming Xu, and Huixiao Hong. 2022. "Deep Learning Models for Predicting Gas Adsorption Capacity of Nanomaterials" Nanomaterials 12, no. 19: 3376. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12193376