Quantum-Chemical Quasi-Docking for Molecular Dynamics Calculations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Main Methods and Software
2.2. Test Set of Protein-Ligand Complexes
3. Results and Discussions
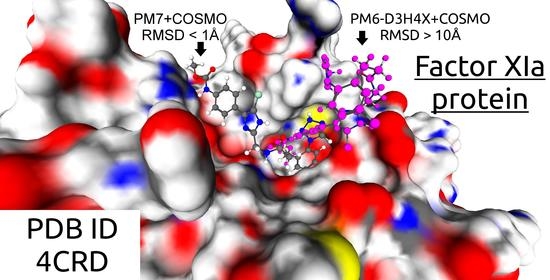
3.1. Ligand Positioning
3.2. Binding Enthalpy
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Director-General’s Remarks at the Media Briefing on 2019-NCoV on 11 February 2020. Available online: Https://www.Who.Int/Dg/Speeches/Detail/Who-Director-General-s-Remarksat-the-Media-Briefing-on-2019-Ncov-on-11-February-2020 (accessed on 11 February 2020).
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Ghahremanpour, M.M.; Tirado-Rives, J.; Deshmukh, M.; Ippolito, J.A.; Zhang, C.-H.; Cabeza de Vaca, I.; Liosi, M.-E.; Anderson, K.S.; Jorgensen, W.L. Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020, 11, 2526–2533. [Google Scholar] [CrossRef]
- Zhang, C.-H.; Stone, E.A.; Deshmukh, M.; Ippolito, J.A.; Ghahremanpour, M.M.; Tirado-Rives, J.; Spasov, K.A.; Zhang, S.; Takeo, Y.; Kudalkar, S.N.; et al. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent. Sci. 2021, 7, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xu, M.; Chen, C.Z.; Guo, H.; Shen, M.; Hu, X.; Shinn, P.; Klumpp-Thomas, C.; Michael, S.G.; Zheng, W. Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-Throughput Screening. ACS Pharmacol. Transl. Sci. 2020, 3, 1008–1016. [Google Scholar] [CrossRef]
- Kuzikov, M.; Costanzi, E.; Reinshagen, J.; Esposito, F.; Vangeel, L.; Wolf, M.; Ellinger, B.; Claussen, C.; Geisslinger, G.; Corona, A.; et al. Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen. ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. [Google Scholar] [CrossRef]
- Sulimov, V.B.; Kutov, D.C.; Sulimov, A.V. Advances in Docking. Curr. Med. Chem. 2019, 26, 7555–7580. [Google Scholar] [CrossRef] [PubMed]
- Sulimov, A.V.; Kutov, D.C.; Sulimov, V.B. Supercomputer Docking. Supercomput. Front. Innov. 2019, 6, 26–50. [Google Scholar] [CrossRef]
- Sulimov, V.B.; Kutov, D.C.; Taschilova, A.S.; Ilin, I.S.; Tyrtyshnikov, E.E.; Sulimov, A. V Docking Paradigm in Drug Design. Curr. Top. Med. Chem. 2021, 21, 507–546. [Google Scholar] [CrossRef]
- Sulimov, A.V.; Kutov, D.C.; Katkova, E.V.; Kondakova, O.A.; Sulimov, V.B. Search for Approaches to Improving the Calculation Accuracy of the Protein-Ligand Binding Energy by Docking. Russ. Chem. Bull. 2017, 66, 1913–1924. [Google Scholar] [CrossRef]
- Sulimov, A.V.; Kutov, D.C.; Katkova, E.V.; Sulimov, V.B. Combined Docking with Classical Force Field and Quantum Chemical Semiempirical Method PM7. Adv. Bioinform. 2017, 2017, 7167691. [Google Scholar] [CrossRef] [Green Version]
- Sulimov, A.V.; Kutov, D.C.; Katkova, E.V.; Ilin, I.S.; Sulimov, V.B. New Generation of Docking Programs: Supercomputer Validation of Force Fields and Quantum-Chemical Methods for Docking. J. Mol. Graph. Model. 2017, 78, 139–147. [Google Scholar] [CrossRef]
- Sulimov, A.V.; Kutov, D.C.; Gribkova, A.K.; Ilin, I.S.; Tashchilova, A.S.; Sulimov, V.B. Search for Approaches to Supercomputer Quantum-Chemical Docking. In Supercomputing. RuSCDays 2019; Communications in Computer and Information Science; Voevodin, V., Sobolev, S., Eds.; Springer: Cham, Switzerland, 2019; Volume 1129, pp. 363–378. [Google Scholar] [CrossRef]
- Řezáč, J.; Hobza, P. A Halogen-Bonding Correction for the Semiempirical PM6 Method. Chem. Phys. Lett. 2011, 506, 286–289. [Google Scholar] [CrossRef]
- Řezáč, J.; Hobza, P. Advanced Corrections of Hydrogen Bonding and Dispersion for Semiempirical Quantum Mechanical Methods. J. Chem. Theory Comput. 2012, 8, 141–151. [Google Scholar] [CrossRef]
- Stewart, J.J. Optimization of Parameters for Semiempirical Methods VI: More Modifications to the NDDO Approximations and Re-Optimization of Parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Hostaš, J.; Řezáč, J.; Hobza, P. On the Performance of the Semiempirical Quantum Mechanical PM6 and PM7 Methods for Noncovalent Interactions. Chem. Phys. Lett. 2013, 568–569, 161–166. [Google Scholar] [CrossRef]
- Oferkin, I.V.; Katkova, E.V.; Sulimov, A.V.; Kutov, D.C.; Sobolev, S.I.; Voevodin, V.V.; Sulimov, V.B. Evaluation of Docking Target Functions by the Comprehensive Investigation of Protein-Ligand Energy Minima. Adv. Bioinform. 2015, 2015, 126858. [Google Scholar] [CrossRef] [Green Version]
- Oferkin, I.V.; Zheltkov, D.A.; Tyrtyshnikov, E.E.; Sulimov, A.V.; Kutov, D.C.; Sulimov, V.B. Evaluation of the Docking Algorithm Based on Tensor Train Global Optimization. Bull. S. Ural State Univ. Ser. Math. Model. Program. Comput. Softw. 2015, 8, 83–99. [Google Scholar] [CrossRef]
- Sulimov, A.V.; Kutov, D.C.; Sulimov, V.B. Parallel Supercomputer Docking Program of the New Generation: Finding Low Energy Minima Spectrum. In Supercomputing. RuSCDays 2018; Communications in Computer and Information Science; Voevodin, V., Sobolev, S., Eds.; Springer: Cham, Switzerland, 2019; Volume 965, pp. 314–330. [Google Scholar] [CrossRef]
- Kutov, D.C.; Sulimov, A.V.; Sulimov, V.B. Supercomputer Docking: Investigation of Low Energy Minima of Protein-Ligand Complexes. Supercomput. Front. Innov. 2018, 5, 134–137. [Google Scholar] [CrossRef]
- Byrd, R.; Lu, P.; Nocedal, J.; Zhu, C. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM J. Sci. Comput. 1995, 16, 1190–1208. [Google Scholar] [CrossRef]
- Zhu, C.; Byrd, R.H.; Lu, P.; Nocedal, J. Algorithm 778: L-BFGS-B: Fortran Subroutines for Large-Scale Bound-Constrained Optimization. ACM Trans. Math. Softw. 1997, 23, 550–560. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. J. Comput. Chem. 1996, 17, 490–641. [Google Scholar] [CrossRef]
- Romanov, A.N.; Jabin, S.N.; Martynov, Y.B.; Sulimov, A.V.; Grigoriev, F.V.; Sulimov, V.B. Surface Generalized Born Method: A Simple, Fast, and Precise Implicit Solvent Model beyond the Coulomb Approximation. J. Phys. Chem. A 2004, 108, 9323–9327. [Google Scholar] [CrossRef]
- Sulimov, A.V.; Kutov, D.K.; Ilin, I.S.; Sulimov, V.B. Docking with Combined Use of a Force Field and a Quantum-Chemical Method. Biomeditsinskaya Khimiya 2019, 65, 80–85. [Google Scholar] [CrossRef] [Green Version]
- Stewart, J.J.P. Stewart Computational Chemistry. MOPAC2016. Available online: http://openmopac.net/MOPAC2016.html (accessed on 30 July 2020).
- Klamt, A.; Schuurmann, G. COSMO: A New Approach to Dielectric Screening in Solvents with Explicit Expressions for the Screening Energy and Its Gradient. J. Chem. Soc. Perkin Trans. 1993, 2, 799–805. [Google Scholar] [CrossRef]
- Kříž, K.; Řezáč, J. Reparametrization of the COSMO Solvent Model for Semiempirical Methods PM6 and PM7. J. Chem. Inf. Model. 2019, 59, 229–235. [Google Scholar] [CrossRef]
- Basilevsky, M.V.; Leontyev, I.V.; Luschekina, S.V.; Kondakova, O.A.; Sulimov, V.B. Computation of Hydration Free Energies of Organic Solutes with an Implicit Water Model. J. Comput. Chem. 2006, 27, 552–570. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Application of Localized Molecular Orbitals to the Solution of Semiempirical Self-Consistent Field Equations. Int. J. Quantum Chem. 1996, 58, 133–146. [Google Scholar] [CrossRef]
- Sulimov, V.B.; Ilin, I.S.; Kutov, D.C.; Sulimov, A.V. Development of Docking Programs for Lomonosov Supercomputer. J. Turkish Chem. Soc. Sect. A Chem. 2020, 7, 259–276. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Kutov, D.C.; Katkova, E.V.; Kondakova, O.A.; Sulimov, A.V.; Sulimov, V.B. Influence of the Method of Hydrogen Atoms Incorporation into the Target Protein on the Protein-Ligand Binding Energy. Bull. S. Ural State Univ. Ser. Math. Model. Program. Comput. Softw. 2017, 10, 94–107. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kříž, K.; Řezáč, J. Benchmarking of Semiempirical Quantum-Mechanical Methods on Systems Relevant to Computer-Aided Drug Design. J. Chem. Inf. Model. 2020, 60, 1453–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voevodin, V.V.; Antonov, A.S.; Nikitenko, D.A.; Shvets, P.A.; Sobolev, S.I.; Sidorov, I.Y.; Stefanov, K.S.; Voevodin, V.V.; Zhumatiy, S.A. Supercomputer Lomonosov-2: Large Scale, Deep Monitoring and Fine Analytics for the User Community. Supercomput. Front. Innov. 2019, 6, 4–11. [Google Scholar] [CrossRef] [Green Version]
| Protein | PDB ID | Res, Å | NP | NL | Ntors |
|---|---|---|---|---|---|
| GNB/LNB-binding protein | 2Z8D | 1.9 | 5897 | 51 | 6 |
| 2Z8E | 2.0 | 5897 | 51 | 6 | |
| α-fucosidase | 2XII | 1.8 | 7042 | 51 | 4 |
| KIV-10 module of Apo (a) | 3KIV | 1.8 | 1206 | 20 | 5 |
| BET protein | 4MR5 | 1.6 | 1860 | 42 | 3 |
| 4MR6 | 1.7 | 1860 | 49 | 6 | |
| CRP | 1HW5 | 1.8 | 3284 | 33 | 1 |
| Trypsin | 1C5P | 1.4 | 3220 | 18 | 1 |
| 1K1J | 2.2 | 3220 | 68 | 10 | |
| 2ZDM | 1.9 | 3220 | 59 | 9 | |
| 2ZDN | 2.0 | 3220 | 58 | 9 | |
| 2ZFS | 1.5 | 3220 | 64 | 9 | |
| YKL-39 | 4P8V | 1.6 | 5741 | 57 | 8 |
| Factor Xia | 4CRC | 1.6 | 3711 | 60 | 11 |
| 4CRD | 2.1 | 3692 | 57 | 11 | |
| EngF | 1J84 | 2.0 | 2642 | 87 | 10 |
| Mp1p-LBD2 | 5CSD | 1.5 | 2407 | 53 | 14 |
| HIV-1 protease | 1MRX | 2.0 | 3140 | 74 | 11 |
| 1MSM | 2.0 | 3138 | 78 | 12 | |
| 2PYM | 1.9 | 3100 | 86 | 12 | |
| 2PYN | 1.9 | 3116 | 86 | 12 | |
| 3KDB | 1.7 | 3138 | 86 | 13 | |
| 3NU3 | 1.0 | 3134 | 70 | 13 | |
| 4LL3 | 2.0 | 3134 | 75 | 13 | |
| Renin | 2IKO | 1.9 | 5144 | 46 | 5 |
| PDB ID | Nmin | PM6-D3H4X COSMO | PM7 COSMO | ||
|---|---|---|---|---|---|
| INN | RMSD, Å | INN | RMSD, Å | ||
| 1C5P | 5349 | 1 | 0.44 | 1 | 0.43 |
| 1HW5 | 6848 | 1 | 0.48 | 1 | 0.48 |
| 1J84 | 8192 | 11 | 5.64 | 1 | 1.97 |
| 1K1J | 8101 | 1 | 0.33 | 1 | 0.33 |
| 1MRX | 2627 | 1 | 1.35 | 1 | 0.47 |
| 1MSM | 6030 | 1 | 1.87 | 1 | 1.87 |
| 2IKO | 2622 | 1 | 0.49 | 1 | 0.49 |
| 2PYM | 4340 | 1 | 1.66 | 1 | 1.66 |
| 2PYN | 4953 | 1 | 1.22 | 1 | 1.22 |
| 2XII | 8192 | 1 | 0.58 | 1 | 0.58 |
| 2Z8D | 8192 | 5 | 3.67 | 2 | 3.67 |
| 2Z8E | 8192 | 1 | 1.11 | 1 | 1.11 |
| 2ZDM | 5971 | 1 | 0.91 | 1 | 0.91 |
| 2ZDN | 5645 | 8 | 2.12 | 1 | 0.68 |
| 2ZFS | 5986 | 9 | 2.67 | 2 | 2.67 |
| 3KDB | 4504 | 2 | 2.61 | 1 | 0.96 |
| 3KIV | 5363 | 1 | 0.84 | 1 | 0.75 |
| 3NU3 | 4935 | 1 | 0.44 | 1 | 0.44 |
| 4CRC | 11809 | 2 | 2.67 | 2 | 2.67 |
| 4CRD | 20222 | 233 | 10.81 | 1 | 1 |
| 4LL3 | 5888 | 3 | 11.67 | 4 | 8.38 |
| 4MR5 | 5002 | 2 | 8.05 | 5 | 8.05 |
| 4MR6 | 4313 | 1 | 1.16 | 1 | 1.16 |
| 4P8V | 8193 | 37 | 10.11 | 1 | 0.58 |
| 5CSD | 29528 | 2576 | 10.33 | 993 | 10.33 |
| PDB ID | ΔHexp, kcal/mol | ΔHbind, kcal/mol | |
|---|---|---|---|
| PM6-D3H4X COSMO | PM7 COSMO | ||
| 1C5P | −4.52 | −27.70 | −54.75 |
| 1HW5 | −0.97 | −37.04 | −54.74 |
| 1K1J | −9.46 | −29.62 | −82.71 |
| 1MRX | −2.10 | −9.36 | −54.89 |
| 1MSM | −7.60 | −20.13 | −67.86 |
| 2IKO | −9.50 | −33.81 | −81.19 |
| 2XII | −9.80 | −72.28 | −92.09 |
| 2ZDM | −7.24 | −30.73 | −82.20 |
| 2ZDN | −5.09 | – | −85.08 |
| 3KDB | −1.55 | – | −54.68 |
| 3NU3 | −7.30 | −6.78 | −54.46 |
| 4MR6 | −4.04 | −15.27 | −47.42 |
| R | 0.4 | 0.74 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulimov, A.; Kutov, D.; Ilin, I.; Sulimov, V. Quantum-Chemical Quasi-Docking for Molecular Dynamics Calculations. Nanomaterials 2022, 12, 274. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12020274
Sulimov A, Kutov D, Ilin I, Sulimov V. Quantum-Chemical Quasi-Docking for Molecular Dynamics Calculations. Nanomaterials. 2022; 12(2):274. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12020274
Chicago/Turabian StyleSulimov, Alexey, Danil Kutov, Ivan Ilin, and Vladimir Sulimov. 2022. "Quantum-Chemical Quasi-Docking for Molecular Dynamics Calculations" Nanomaterials 12, no. 2: 274. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12020274