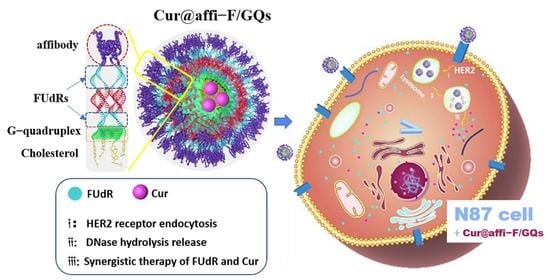
Affibody Modified G-quadruplex DNA Micelles Incorporating Polymeric 5-Fluorodeoxyuridine for Targeted Delivery of Curcumin to Enhance Synergetic Therapy of HER2 Positive Gastric Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Synthesis of DNA Micelles
2.2.1. Synthesis of FUdR Phosphoramidite
2.2.2. Solid-Phase Synthesis of cF10G6-Chl and cF13-NH2
2.2.3. Synthesis, Purification and Characterization of cF13-affibody Conjugate
2.2.4. Self-Assembly of affi-F/GQs
2.2.5. Preparation of Cur@affi-F/GQs
2.3. Characterization
2.3.1. Determination of Critical Micelle Concentration (CMC)
2.3.2. Stability Analysis of affi-F/GQs
2.3.3. Drug Loading Study of Cur@affi-F/GQs
2.3.4. In Vitro Drug Release Study of Cur@affi-F/GQs
2.3.5. Circular Dichroism (CD) Spectroscopy
2.3.6. Atomic force microscopy (AFM)
2.3.7. Transmission Electron Microscopy (TEM)
2.3.8. Dynamic Light Scattering (DLS)
2.4. Cellular Experiments
2.4.1. Cell Lines and Cell Cultures
2.4.2. Cellular Uptake
2.4.3. In Vitro Cytotoxicity
2.4.4. Western Blot Analysis
2.4.5. Caspase Activity Assays
2.5. Statistical Analysis
3. Results and discussion
3.1. Structural Design of affi-F/GQs
3.2. Synthesis and Characterization of cF10G6-Chl and cF13-affibody
3.3. Self-assembly and Characterization of affi-F/GQs
3.4. Determination of Critical Micelle Concentration
3.5. Stability Analysis of affi-F/GQs
3.6. Preparation and Characterization of Cur@affi-F/GQs
3.7. In Vitro Drug Release Behavior
3.8. Cellular Uptake
3.9. Cytotoxicity and Synergistic Effect Studies
3.10. Exploration of Anticancer Mechanism
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| ssDNA | Sequence |
|---|---|
| cF13 | 5’-cagtctgatagcgFFFFFFFFFFFFF-3’ |
| cF10G6 | 5’-cgctatcagactgFFFFFFFFFFGGGGGG-3’ |
| cF13-NH2 | 5’-cagtctgatagcgFFFFFFFFFFFFF-NH2-3’ |
| cF10G6-Chl | 5’-cgctatcagactgFFFFFFFFFFGGGGGG-Cholesterol-3’ |
| cF10T6-Chl | 5’-cgctatcagactgFFFFFFFFFFTTTTTT-Cholesterol-3’ |
| FAM-cF10G6-Chl | 5’-FAM/cgctatcagactgFFFFFFFFFFGGGGGG-Cholesterol-3’ |








References
- Hu, Q.; Sun, W.; Wang, C.; Gu, Z. Recent advances of cocktail chemotherapy by combination drug delivery systems. Adv. Drug Deliv. Rev. 2016, 98, 19–34. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Kohli, M.; Smith, A. Nanoparticles for combination drug therapy. ACS Nano 2013, 7, 9518–9525. [Google Scholar] [CrossRef] [Green Version]
- Tangalos, E.G.; Zarowitz, B.J. Combination drug therapy. J. Am. Med. Dir. Assoc. 2005, 6, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ho, W.; Zhang, X.; Bertrand, N.; Farokhzad, O. Cancer nanomedicine: From targeted delivery to combination therapy. Trends Mol. Med. 2015, 21, 223–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Y.; Wang, R.; Gao, L.; Li, K.; Zhou, X.; Guo, H.; Liu, C.; Han, D.; Tian, J.; Ye, Q.; et al. Sequential co-delivery of miR-21 inhibitor followed by burst release doxorubicin using NIR-responsive hollow gold nanoparticle to enhance anticancer efficacy. J. Controlled. Release 2016, 228, 74–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, P.Y.; Cheng, W.; Hedrick, J.L.; Yang, Y.Y. Co-delivery of drugs and plasmid DNA for cancer therapy. Adv. Drug Deliv. Rev. 2016, 98, 41–63. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Sliwinska, P.; Weiss, A.; Ding, X.; Dyson, P.J.; Bergh, H.V.D.; Griffioen, A.W.; Ho, C.M. Optimization of drug combinations using Feedback System Control. Nat. Protoc. 2016, 11, 302–315. [Google Scholar] [CrossRef]
- Xiao, B.; Han, M.K.; Viennois, E.; Wang, L.; Zhang, M.; Sia, X.; Merlinbc, D. Hyaluronic acid-functionalized polymeric nanoparticles for colon cancer-targeted combination chemotherapy. Nanoscale 2015, 7, 17745–17755. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, B.G.; Vit, F.F.; Carvalho, H.F.; Hanc, S.W.; de la Torre, L.G. Recent advances in co-delivery nanosystems for synergistic action in cancer treatment. J. Mater. Chem. B 2021, 9, 1208–1237. [Google Scholar] [CrossRef]
- Edis, Z.; Wang, J.; Waqas, M.K.; Ijaz, M.; Ijaz, M. Nanocarriers-mediated drug delivery systems for anticancer agents: An overview and perspectives. Int. J. Nanomed. 2021, 16, 1313–1330. [Google Scholar] [CrossRef]
- Su, S.; Kang, P.M. Recent advances in nanocarrier-assisted therapeutics delivery systems. Pharmaceutics 2020, 12, 837. [Google Scholar] [CrossRef]
- Rahdar, A.; Hajinezhad, M.R.; Sargazi, S.; Barani, M.; Karimi, P.; Velasco, B.; Taboada, P.; Pandey, S.; Bameri, Z.; Zarei, S. Pluronic F127/carfilzomib-based nanomicelles as promising nanocarriers: Synthesis, characterization, biological, and in silico evaluations. J. Mol. Liq. 2022, 346, 118271. [Google Scholar] [CrossRef]
- Barani, M.; Sargazi, S.; Hajinezhad, M.R.; Rahdar, A.; Sabir, F.; Pardakhty, A.; Zargari, F.; Anwer, M.K.; Aboudzadeh, M.A. Preparation of pH-responsive vesicular deferasirox: Evidence from in silico, in vitro, and in vivo evaluations. ACS Omega 2021, 6, 24218–24232. [Google Scholar] [CrossRef]
- Nezhadali, A.; Shapouri, M.R.; Amoli-Diva, M. Anti-cancer combination therapy by co-delivery of hydrophilic and hydrophobic using dual temperature and pH-responsive liposomes. Micro Nano. Lett. 2020, 15, 1065–1070. [Google Scholar] [CrossRef]
- Naderinezhad, S.; Amoabediny, G.; Haghiralsadatb, F. Co-delivery of hydrophilic and hydrophobic anticancer drugs using biocompatible pH-sensitive lipid-based nano-carriers for multidrug-resistant cancers. RSC Adv. 2017, 7, 30008–30019. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, J.; Thayumanavan, S. Triblock-diblock composite nanoassemblies with sequentially addressable host–guest properties for hydrophobics and hydrophilics. ACS Macro Lett. 2020, 9, 1019–1023. [Google Scholar] [CrossRef]
- Danafar, H.; Rostamizadeh, K.; Davaran, S.; Hamidi, M. Co-delivery of hydrophilic and hydrophobic drugs by micelles: A new approach using drug conjugated PEG–PCLNanoparticles. Drug Dev. Ind. Pharm. 2017, 43, 1908–1918. [Google Scholar] [CrossRef]
- Li, S.; Gan, Y.; Lin, C.; Lin, K.; Hu, P.; Liu, L.; Yu, S.; Zhao, S.; Shi, J. NIR-/pH-responsive nanocarriers based on mesoporous hollow polydopamine for codelivery of hydrophilic/hydrophobic drugs and photothermal synergetic therapy. ACS Appl. Bio Mater. 2021, 4, 1605–1615. [Google Scholar] [CrossRef]
- Greco, F.; Vicent, M.J. Combination therapy: Opportunities and challenges for polymer-drug conjugates as anticancer nanomedicines. Adv. Drug Deliv. Rev. 2009, 61, 1203–1213. [Google Scholar] [CrossRef]
- Zhang, R.X.; Wong, H.L.; Xue, H.Y.; Eoh, J.Y.; Wu, X.Y. Nanomedicine of synergistic drug combinations for cancer therapy-Strategies and perspectives. J. Control. Release 2016, 240, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Seeman, N.; Sleiman, H. DNA nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. [Google Scholar] [CrossRef]
- Kumar, V.; Palazzolo, S.; Bayda, S.; Corona, G.; Toffoli, G.; Rizzolio, F. DNA Nanotechnology for Cancer Therapy. Theranostics 2016, 6, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Alemdaroglu, F.E.; Herrmann, A. DNA meets synthetic polymers-highly versatile hybrid materials. Org. Biomol. Chem. 2007, 6, 1311–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alemdaroglu, F.E.; Alemdaroglu, N.C.; Langguth, P.; Herrmann, A. DNA block copolymer micelles—A combinatorial tool for cancer nanotechnology. Adv. Mater. 2008, 20, 899–902. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.H.; Park, T.G. Novel polymer-DNA hybrid polymeric micelles composed of hydrophobic poly(d,l-lactic-co-glycolic acid) and hydrophilic oligonucleotides. Bioconjugate Chem. 2001, 12, 917–923. [Google Scholar] [CrossRef]
- Trinh, T.; Chidchob, P.; Bazzi, H.S.; Sleiman, H.F. DNA micelles as nanoreactors: Efficient DNA functionalization with hydrophobic organic molecules. Chem. Commun. 2016, 52, 10914–10917. [Google Scholar] [CrossRef]
- Johnson-Buck, A.; Jiang, S.; Yan, H.; Walter, N.G. DNA-cholesterol barges as programmable membrane-exploring agents. ACS Nano 2014, 8, 5641–5649. [Google Scholar] [CrossRef]
- Chen, T.; Wu, C.S.; Jimenez, E.; Zhu, Z.; Dajac, J.G.; You, M.; Han, D.; Zhang, X.; Tan, W.H. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew. Chem. Int. Ed. Engl. 2013, 52, 2012–2016. [Google Scholar] [CrossRef] [Green Version]
- Lyu, Y.; Guo, Y.; Cai, R.; Peng, R.; Hong, C.; Chen, X.; Hou, W.; Li, X.; Tan, J.; Zou, Y.; et al. Spherically directed synthesis and enhanced cellular internalization of metal-crosslinked DNA micelles. Chem 2019, 5, 913–928. [Google Scholar] [CrossRef] [Green Version]
- Ilkhani, H.; Hughes, T.; Li, J.; Zhong, C.J.; Hepel, M. Nanostructured SERS-electrochemical biosensors for testing of anticancer drug interactions with DNA. Biosens. Bioelectron. 2016, 80, 257–264. [Google Scholar] [CrossRef]
- Kastantin, M.; Missirlis, D.; Black, M.; Ananthanarayanan, B.; Peters, D.; Tirrell, M. Thermodynamic and kinetic stability of DSPE-PEG(2000) micelles in the presence of bovine serum albumin. J. Phys. Chem. B 2010, 114, 12632. [Google Scholar] [CrossRef] [PubMed]
- Castelletto, V.; Krysmann, M.; Kelarakis, A.; Jauregi, P. Complex formation of bovine serum albumin with a poly(ethylene glycol) lipid conjugate. Biomacromolecules 2007, 8, 2244–2249. [Google Scholar] [CrossRef] [PubMed]
- Brune, V.; Gissot, A.; Delzor, R.; Barthelemy, P. Controlling G-quadruplex formation via lipid modification of oligonucleotide sequences. Chem. Commun. 2017, 53, 11560–11563. [Google Scholar] [CrossRef] [Green Version]
- Cozzoli, L.; Gjonaj, L.; Stuart, M.C.A.; Poolman, B.; Roelfes, G. Responsive DNA G-quadruplex micelles. Chem. Commun. 2018, 54, 260–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, J.T. G-quartets 40 years later: From 5’-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004, 43, 668–698. [Google Scholar] [CrossRef] [PubMed]
- Wilner, S.E.; Sparks, S.E.; Cowburn, D.; Girvin, M.E.; Levy, M. Controlling lipid micelle stability using oligonucleotide headgroups. J. Am. Chem. Soc. 2015, 137, 2171–2174. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Liu, X.; Bai, H.; Wang, R.; Tan, J.; Peng, X.; Tan, W. Engineering Stability-Tunable DNA Micelles Using Photocontrollable Dissociation of an Intermolecular G-Quadruplex. ACS Nano 2017, 11, 12087–12093. [Google Scholar] [CrossRef]
- Ilkhani, H.; Zhong, C.-J.; Hepel, M. Magneto-plasmonic nanoparticle grid biosensor with enhanced raman scattering and electrochemical transduction for the development of nanocarriers for targeted delivery of protected anticancer drugs. Nanomaterials 2021, 11, 1326. [Google Scholar] [CrossRef]
- Ilkhani, H.; Sarparast, M.; Noori, A.; Bathaie, S.Z.; Mousavi, M.F. Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker, using gold nanoparticles as a signaling probe. Biosens. Bioelectron. 2015, 74, 491–497. [Google Scholar] [CrossRef]
- Bang, J.; Park, H.; Choi, W.I.; Sung, D.; Lee, J.H.; Lee, K.Y.; Kim, S. Sensitive detection of dengue virus NS1 by highly stable affibody-functionalized gold nanoparticles. New J. Chem. 2018, 42, 12607–12614. [Google Scholar] [CrossRef]
- Liu, J.; Cui, D.; Jiang, Y.; Li, Y.; Liu, Z.; Tao, L.; Zhao, Q.; Diao, A. Selection and characterization of a novel affibody peptide and its application in a two-site ELISA for the detection of cancer biomarker alpha-fetoprotein. Int. J. Biol. Macromol. 2021, 166, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Ilkhani, H.; Ravalli, A.; Marrazza, G. Design of an affibody-based recognition strategy for human epidermal growth factor receptor 2 (HER2) detection by electrochemical biosensors. Chemosensors 2016, 4, 23. [Google Scholar] [CrossRef]
- Zhang, C.; Han, M.; Zhang, F.; Yang, X.; Du, J.; Zhang, H.; Li, W.; Chen, S. Enhancing antitumor efficacy of nucleoside analog 5-fluorodeoxyuridine on HER2-overexpressing breast cancer by affibody-engineered DNA nanoparticle. Int. J. Nanomed. 2020, 15, 885–900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yin, J.; Zhang, C.; Han, M.; Wang, X.; Fu, S.; Du, J.; Zhang, H.; Li, W. Affibody-conjugated RALA polymers delivering oligomeric 5-fluorodeoxyuridine for targeted therapy of HER2 overexpressing gastric cancer. Macromol. Biosci. 2020, 20, 2000083. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, F.; Han, M.; Wang, X.; Du, J.; Zhang, H.; Li, W. Co-delivery of 5-fluorodeoxyuridine and doxorubicin via gold nanoparticle equipped with affibody-DNA hybrid strands for targeted synergistic chemotherapy of HER2 overexpressing breast cancer. Sci. Rep. 2020, 10, 22015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lai, Z.L.; Chen, H.F.; Zhang, M.; Wang, A.; Jia, T.; Sun, Q.W.; Zhu, M.X.; Chen, F.X. Curcumin synergizes with 5-fluorouracil by impairing AMPK/ULK1-dependent autophagy, AKT activity and enhancing apoptosis in colon cancer cells with tumor growth inhibition in xenograft mice. J. Exp. Clin. Cancer Res. 2017, 36, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afsharmoghadam, N.; Haghighatian, Z.; Mazdak, H.; Mirkheshti, N.; Mehrabi Koushki, R.; Alavi, S.A. Concentration-dependent effects of curcumin on 5-fluorouracil efficacy in bladder cancer cells. Asian Pac. J. Cancer Prev. 2017, 18, 3225–3230. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.E.; Orlando, R.A. Curcumin reduces cytotoxicity of 5-Fluorouracil treatment in human breast cancer cells. J. Med. Food 2015, 18, 497–502. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Rahdar, A.; Hajinezhad, M.R.; Sargazi, S.; Zaboli, M.; Barani, M.; Baino, F.; Bilal, M.; Sanchooli, E. Biochemical, ameliorative and cytotoxic effects of newly synthesized curcumin microemulsions: Evidence from in vitro and in vivo studies. Nanomaterials 2021, 11, 817. [Google Scholar] [CrossRef]
- Naksuriya, O.; Okonogi, S.; Schiffelers, R.M.; Hennink, W.E. Curcumin nanoformulations: A review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials 2014, 35, 3365–3383. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Yang, P.; Cao, S.; Zhao, L. The combination of curcumin and 5-fluorouracil in cancer therapy. Arch. Pharm. Res. 2018, 41, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Marasco, C.J.J.; Sufrin, J.R.A. A convenient method for the direct incorporation of 5-fluoro-2′-deoxycytidine into oligodeoxynucleotides. J. Org. Chem. 1992, 57, 6363–6365. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Gray, R.D.; Chaires, J.B. Characterization of quadruplex DNA structure by circular dichroism. Curr. Protoc. Nucleic. Acid Chem. 2017, 68, 17.8.1–17.8.16. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Trotta, R.; Pieraccini, S.; Tito, S.D.; Perone, R.; Randazzo, A.; Spada, G.P. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010, 8, 2683–2692. [Google Scholar] [CrossRef]
- Zana, R. Fluorescence studies of amphiphilic block copolymers in solution. Amphiphilic Block Copolym. 2000, 244, 221–252. [Google Scholar] [CrossRef]
- Miller, T.; Rachel, R.; Besheer, A.; Uezguen, S.; Weigandt, M.; Goepferichet, A. Comparative investigations on in vitro serum stability of polymeric micelle formulations. Pharm. Res. 2012, 29, 448–459. [Google Scholar] [CrossRef]
- Breitenkamp, K. Polymeric micelles for drug delivery. Curr. Pharm. Des. 2006, 12, 4669–4684. [Google Scholar] [CrossRef]
- Hiremath, C.G.; Kariduraganavar, M.Y.; Hiremath, M.B. Synergistic delivery of 5-fluorouracil and curcumin using human serum albumin-coated iron oxide nanoparticles by folic acid targeting. Prog. Biomater. 2018, 7, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Anitha, A.; Deepa, N.; Chennazhi, K.P.; Lakshmanan, V.K.; Jayakumar, R. Combinatorial anticancer effects of curcumin and 5-fluorouracil loaded thiolated chitosan nanoparticles towards colon cancer treatment. Biochim. Biophys. Acta 2014, 1840, 2730–2743. [Google Scholar] [CrossRef]
- Srivastava, S.; Mohammad, S.; Pant, A.B.; Mishra, P.R.; Pandey, G.; Gupta, S.; Farooqui, S. Co-delivery of 5-fluorouracil and curcumin nanohybrid formulations for improved chemotherapy against oral squamous cell carcinoma. J. Maxillofac. Oral Surg. 2018, 17, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.-J.; Jin, Y.; Xie, M.Q.; Ye, Y.-J.; Qin, D.-D.; Lou, K.-Y.; Chen, Y.-Z.; Gao, F. Preparation and characterization of mPEG grafted chitosan micelles as 5-fluorouracil carriers for effective anti-tumor activity. Chin. Chem. Lett. 2014, 25, 1435–1440. [Google Scholar] [CrossRef]
- Gu, C.; Le, V.; Lang, M.; Liu, J. Preparation of polysaccharide derivates chitosan-graft-poly(ε-caprolactone) amphiphilic copolymer micelles for 5-fluorouracil drug delivery. Colloids Surf. B 2014, 116, 745–750. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Tang, J.N.; Xie, H.X.; Du, Y.A.; Huang, L.; Yu, P.F.; Cheng, X.D. 5-Fluorouracil chemotherapy of gastric cancer generates residual cells with properties of cancer stem cells. Int. J. Biol. Sci. 2015, 11, 284–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.Y.; Koh, C.E.; Bu, Z.D.; Wu, A.W.; Zhang, L.H.; Wu, X.J.; Wu, Q.; Zong, X.L.; Ren, H.; Tang, L.; et al. Neoadjuvant chemotherapy with FOLFOX: Improved outcomes in Chinese patients with locally advanced gastric cancer. J. Surg. Oncol. 2012, 105, 793–799. [Google Scholar] [CrossRef]
- Ashton, J.C. Drug combination studies and their synergy quantification using the Chou–Talalay method–letter. Cancer Res. 2015, 75, 2400. [Google Scholar] [CrossRef] [Green Version]
- Sato, A.; Hiramoto, A.; Uchikubo, Y.; Miyazaki, E.; Satake, A.; Naito, T.; Hiraoka, O.; Miyake, T.; Kim, H.; Wataya, Y. Gene expression profiles of necrosis and apoptosis induced by 5–fluoro–2′–deoxyuridine. Genomics 2008, 92, 9–17. [Google Scholar] [CrossRef] [Green Version]
- García-Saez, A. The secrets of the Bcl-l2 family. Cell Death Differ. 2012, 19, 1733. [Google Scholar] [CrossRef] [Green Version]
- Earnshaw, W.C.; Martins, L.M.; Kaufmann, S.H. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999, 68, 383–424. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Fu, S.; Zhang, F.; Han, M.; Wang, X.; Du, J.; Zhang, H.; Li, W. Affibody Modified G-quadruplex DNA Micelles Incorporating Polymeric 5-Fluorodeoxyuridine for Targeted Delivery of Curcumin to Enhance Synergetic Therapy of HER2 Positive Gastric Cancer. Nanomaterials 2022, 12, 696. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12040696
Zhang C, Fu S, Zhang F, Han M, Wang X, Du J, Zhang H, Li W. Affibody Modified G-quadruplex DNA Micelles Incorporating Polymeric 5-Fluorodeoxyuridine for Targeted Delivery of Curcumin to Enhance Synergetic Therapy of HER2 Positive Gastric Cancer. Nanomaterials. 2022; 12(4):696. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12040696
Chicago/Turabian StyleZhang, Chao, Shuangqing Fu, Fanghua Zhang, Mengnan Han, Xuming Wang, Jie Du, Honglei Zhang, and Wei Li. 2022. "Affibody Modified G-quadruplex DNA Micelles Incorporating Polymeric 5-Fluorodeoxyuridine for Targeted Delivery of Curcumin to Enhance Synergetic Therapy of HER2 Positive Gastric Cancer" Nanomaterials 12, no. 4: 696. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12040696