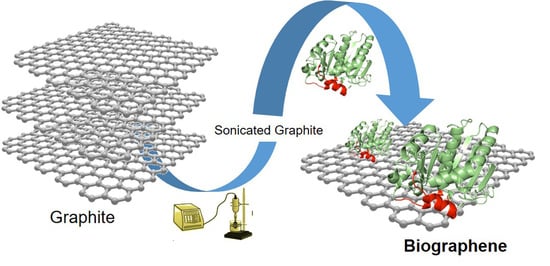
Efficient Production of Multi-Layer Graphene from Graphite Flakes in Water by Lipase-Graphene Sheets Conjugation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterization Techniques
2.3. Ultrasonication of Graphite Flakes
2.3.1. Method 1: Simple Sonication of Graphite Flakes
2.3.2. Method 2: Double Sonication of Graphite Flakes
2.4. Exfoliation of Graphite Flakes Using Lipase
2.4.1. Method 3: Double Sonication of Graphite Flakes + Selective Lipase Adsorption
2.4.2. Method 4: Double Sonication of Graphite Flakes in the Presence of Lipase
2.5. Hydrolytic Activity Assay of pNPP
3. Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Geim, A.K. Graphene: Status and prospects. Science 2009, 324, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Kalantar-Zadeh, K.; Kis, A.; Coleman, J.N.; Strano, M.S. Electronics and optoelectronics of two-dimensional transition metal dichalcogenides. Nat. Nanotechnol. 2012, 7, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Peydayesh, M.; Mezzenga, R. Sustainable technologies for water purification from heavy metals: Review and analysis. Chem. Soc. Rev. 2019, 48, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Novoselov, K.S.; Fal’Ko, V.I.; Colombo, L.; Gellert, P.R.; Schwab, M.G.; Kim, K.A. A roadmap for graphene. Nature 2012, 490, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Schwierz, F. Graphene transistors. Nat. Nanotechnol. 2010, 5, 487–496. [Google Scholar]
- Balandin, A.A. Thermal properties of graphene and nanostructured carbon materials. Nat.Mat. 2011, 10, 569–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodenough, J.B.; Park, K.-S. The Li-ion rechargeable battery: A perspective. J.Am.Chem.Soc. 2013, 135, 1167–1176. [Google Scholar] [CrossRef]
- Bonaccorso, F.; Colombo, L.; Yu, G.; Stoller, M.; Tozzini, V.; Ferrari, A.C.; Ruoff, R.S.; Pellegrini, V. Graphene, related two-dimensional crystals, and hybrid systems for energy conversion and storage. Science 2015, 347, 1246501. [Google Scholar] [CrossRef]
- Yang, K.; Feng, L.; Shi, X.; Liu, Z. Nano-graphene in biomedicine: Theranostic applications. Chem. Soc. Rev. 2013, 42, 530–547. [Google Scholar] [CrossRef]
- Mak, K.F.; Shan, J. Photonics and optoelectronics of 2D semiconductor transition metal dichalcogenides. Nat. Photonics 2016, 10, 216–226. [Google Scholar] [CrossRef]
- Yao, S.; Zhu, Y. Nanomaterial-enabled stretchable conductors: Strategies, materials and devices. Adv. Mat. 2015, 27, 1480–1511. [Google Scholar] [CrossRef] [PubMed]
- Manzeli, S.; Ovchinnikov, D.; Pasquier, D.; Yazyev, O.V.; Kis, A. 2D transition metal dichalcogenides. Nat. Rev. Mat. 2017, 2, 17033. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, H.; Liu, Y.; Li, J. Graphene and its derivatives for the development of solar cells, photoelectrochemical and photocatalytic applications. Energy Environ. Sci. 2013, 6, 1362–1387. [Google Scholar] [CrossRef]
- Novoselov, K.S.; McCann, E.; Morozov, S.V.; Falko, V.I.; Katsnelson, M.I.; Zeitler, U.; Jiang, D.; Schedin, F.; Geim, A.K. Unconventional quantum Hall effect and Berry’s phase of 2π in bilayer graphene. Nat. Phys. 2006, 2, 177–180. [Google Scholar]
- Zhou, X.; Liu, Z. A scalable, solution-phase processing route to graphene oxide and graphene ultralarge sheets. Chem. Commun. 2010, 46, 2611–2613. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, V.; Otyepka, M.; Bourlinos, A.B.; Chandra, V.; Kim, N.; Kemp, K.C.; Hobza, P.; Zboril, R.; Kim, K.S. Functionalization of graphene: Covalent and non-covalent approaches, derivatives and applications. Chem. Rev. 2012, 112, 6156–6214. [Google Scholar]
- Park, S.; Ruoff, R.S. Chemical methods for the production of graphenes. Nat. Nanotechnol. 2009, 4, 217–224. [Google Scholar]
- Tung, V.C.; Allen, M.J.; Yang, Y.; Kaner, R.B. High-throughput solution processing of large-scale graphene. Nat. Nanotechnol. 2009, 4, 25–29. [Google Scholar] [CrossRef]
- Gravagnuolo, A.M.; Morales-Narváez, E.; Longobardi, S.; da Silva, E.T.; Giardina, P.; Merkoçi, A. In Situ Production of Biofunctionalized Few-Layer Defect-Free Microsheets of Graphene. Adv. Funct. Mater. 2015, 25, 2771–2779. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.J.; Kim, K.K.; Benayad, A.; Yoon, S.M.; Park, H.K.; Jung, I.S.; Jim, M.H.; Jeong, H.K.; Kim, J.M.; Choi, J.Y. Efficient Reduction of Graphite Oxide by Sodium Borohydride and Its Effect on Electrical Conductance. Adv. Funct. Mater. 2009, 19, 1987–1992. [Google Scholar] [CrossRef]
- Wei, P.; Gan, T.; Wu, K. N-methyl-2-pyrrolidone exfoliated graphene as highly sensitive analytical platform for carbendazim. Sens. Actuators B Chem. 2018, 274, 551–559. [Google Scholar] [CrossRef]
- León, V.; Rodriguez, A.M.; Prieto, P.; Prato, M.; Vázquez, E. Exfoliation of Graphite with Triazine Derivatives under Ball-Milling Conditions: Preparation of Few-Layer Graphene via Selective Noncovalent Interactions. ACS Nano 2014, 8, 563–571. [Google Scholar] [CrossRef]
- Zhao, S.; Xie, S.; Zhao, Z.; Zhang, J.; Li, L.; Xin, Z. Efficiency Production of Graphene by Tannic Acid-Assisted Exfoliation of Graphite in Water. ACS Sust. Chem. Eng. 2018, 6, 7652–7661. [Google Scholar] [CrossRef]
- Zhuo, H.; Zhang, X.; Wang, L.; Lu, Q.; Kaplan, D.L. Sonication Exfoliation of Defect-Free Graphene in Aqueous Silk Nanofiber Solutions. ACS Sust. Chem. Eng. 2018, 6, 12261–12267. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Chen, T.; Liu, H.-H.; Wang, X.-C.; Zhang, X.-X. Direct Liquid Phase Exfoliation of Graphite to Produce Few-Layer Graphene by Microfluidization. J. Nanosci. Nanotechnol. 2019, 9, 2078–2086. [Google Scholar] [CrossRef] [PubMed]
- Salunke, B.K.; Kim, B.S. Facile synthesis of graphene using a biological method. RSC Adv. 2016, 6, 17158. [Google Scholar] [CrossRef]
- Pattammattel., A.; Kumar, C.V. Kitchen Chemistry 101: Multigram Production of High Quality Biographene in a Blender with Edible Proteins. Adv. Funct. Mater. 2015, 25, 7088–7098. [Google Scholar] [CrossRef]
- Laaksonen, P.; Kainlauri, M.; Laaksonen, T.; Shchepetov, A.; Jiang, H.; Ahopelto, J.; Linder, M.B. Interfacial engineering by proteins: Exfoliation and functionalization of graphene by hydrophobins. Angew. Chem. Int. Ed 2010, 49, 4946–4949. [Google Scholar] [CrossRef] [PubMed]
- Babu, D.B.; Ramesha, K. Melamine assisted liquid exfoliation approach for the synthesis of nitrogen doped graphene-like carbon nano sheets from bio-waste bagasse material and its application towards high areal density Li-S batteries. Carbon 2019, 144, 582–590. [Google Scholar] [CrossRef]
- Linder, M.B.; Szilvay, G.; Nakari-Setälä, T.; Penttilä, M. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 877–896. [Google Scholar] [CrossRef] [PubMed]
- Van Tilbeurgh, H.; Egloff, M.P.; Martinez, C.; Rugani, N.; Verger, R.; Cambillau, C. Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography. Nature. 1993, 362, 814–820. [Google Scholar] [CrossRef]
- Marciello, M.; Filice, M.; Palomo, J.M. Different strategies to enhance activities of lipase catalysts. Catal Sci. Technol. 2012, 2, 1531–1543. [Google Scholar] [CrossRef]
- Palomo, J.M.; Munoz, G.; Fernandez-Lorente, G.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Interfacial adsorption of lipases on very hydrophobic support (octadecyl—Sepabeads): Immobilization, hyperactivation and stabilization of the open form of lipases. J. Mol. Catal. B: Enzym. 2002, 19–20, 279–286. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fuentes, M.; Fernández-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernández-Lafuente, R. General trend of Lipase to auto-assemble giving bi-molecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Guisan, J.M.; Fernandez-Lafuente, R. Specificity enhancement towards hydrophobic substrates by immobilization of lipases by interfacial activation on hydrophobic supports. Enzyme Microb. Technol. 2007, 41, 565–569. [Google Scholar]
- Hermanová, S.; Zarevúcká, M.; Bouša, D.; Mikulics, M.; Sofer, Z. Lipase enzymes on graphene oxide support for high-efficiency Biocatalysis. App. Mat. Today 2016, 5, 200–208. [Google Scholar]
- Mathesh, M.; Luan, B.; Akanbi, T.O.; Weber, J.K.; Liu, J.; Barrow, C.J.; Zhou, R.; Yang, W. Opening Lids: Modulation of Lipase Immobilization by Graphene Oxides. ACS Catal. 2016, 6, 4760–4768. [Google Scholar] [CrossRef]
- Shih, C.-J.; Strano, M.S.; Blankschtein, D. Wetting translucency of graphene. Nature Mat. 2013, 12, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Graf, D.; Molitor, F.; Ensslin, K.; Stampfer, C.; Jungen, A.; Hierold, C.; Wirtz, L. Spatially resolved Raman spectroscopy of single- and few-layer graphene. Nano Lett. 2007, 7, 238–242. [Google Scholar] [CrossRef]
- May, P.; Lazzeri, M.; Venezuela, P.; Herziger, F.; Callsen, G.; Reparaz, J.S.; Hoffmann, A.; Mauri, F.; Maultszch, J. Signature of the two-dimensional phonon dispersion in graphene probed by double-resonantRaman scattering. Phys. Rev. B 2013, 87, 075402/1–6. [Google Scholar] [CrossRef]









© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Losada-Garcia, N.; Berenguer-Murcia, A.; Cazorla-Amorós, D.; Palomo, J.M. Efficient Production of Multi-Layer Graphene from Graphite Flakes in Water by Lipase-Graphene Sheets Conjugation. Nanomaterials 2019, 9, 1344. https://0-doi-org.brum.beds.ac.uk/10.3390/nano9091344
Losada-Garcia N, Berenguer-Murcia A, Cazorla-Amorós D, Palomo JM. Efficient Production of Multi-Layer Graphene from Graphite Flakes in Water by Lipase-Graphene Sheets Conjugation. Nanomaterials. 2019; 9(9):1344. https://0-doi-org.brum.beds.ac.uk/10.3390/nano9091344
Chicago/Turabian StyleLosada-Garcia, Noelia, Angel Berenguer-Murcia, Diego Cazorla-Amorós, and Jose M. Palomo. 2019. "Efficient Production of Multi-Layer Graphene from Graphite Flakes in Water by Lipase-Graphene Sheets Conjugation" Nanomaterials 9, no. 9: 1344. https://0-doi-org.brum.beds.ac.uk/10.3390/nano9091344