A Paper-Based Colorimetric Aptasensor for the Detection of Gentamicin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Gold Nanoparticle Synthesis and Characterization
2.3. Preparation of Aptamer and Modified AuNPs
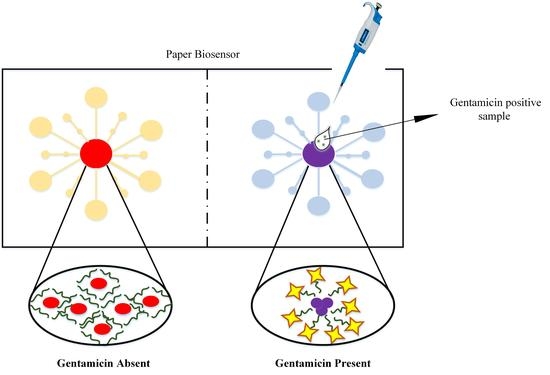
2.4. Preparation and Detection of Gentamicin on Paper-Substrate
2.5. Real Sample Detection of Gentamicin
2.6. Imaging and Analysis of Paper Sensor
3. Results
3.1. Gold Nanoparticles in Optical Detection
3.2. Optimization of Sensing Parameters
4. Discussion
4.1. Sensor Validation
4.2. Gentamicin Paper Assay
4.3. Selectivity Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. About Antibiotic Resistance. Antibiotic/Antimicrobial Resistance. Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 13 April 2020).
- McKellar, Q. Antimicrobial resistance: A veterinary perspective. BMJ 1998, 317, 610–611. [Google Scholar] [CrossRef] [PubMed]
- Wegener, H.C. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 2003, 6, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Brady, M.S.; Katz, S.E. Antibiotic/Antimicrobial Residues in Milk. J. Food Prot. 1988, 51, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Sukul, P.; Spiteller, M. Fluoroquinolone Antibiotics in the Environment. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2007; Volume 191, pp. 131–162. [Google Scholar] [CrossRef]
- Sachi, S.; Ferdous, J.; Sikder, M.H.; Hussani, S.M.A.K. Antibiotic residues in milk: Past, present, and future. J. Adv. Veter. Anim. Res. 2019, 6, 315–332. [Google Scholar] [CrossRef]
- Seymour, E.; Jones, G.; McGilliard, M. Persistence of Residues in Milk Following Antibiotic Treatment of Dairy Cattle. J. Dairy Sci. 1988, 71, 2292–2296. [Google Scholar] [CrossRef]
- Haug, A.; Høstmark, A.T.; Harstad, O.M. Bovine milk in human nutrition—A review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Statista. Global Consumption of Milk per Year by Country. 2019. Available online: https://0-www-statista-com.brum.beds.ac.uk/statistics/272003/global-annual-consumption-of-milk-by-region/ (accessed on 13 April 2020).
- FAO; OMS. CODEX: Maximun Residue Limits (MRLs) and Risk Management Recommendations (RMRs) for Residues of Veterinary Drugs in Foods; CAC/MRL 2-2015; FAO: Rome, Italy, 2015; p. 22. [Google Scholar]
- Rosenkrantz, B.E.; Greco, J.R.; Hoogerheide, J.G.; Oden, E.M. Gentamicin Sulfate. Anal. Profiles Drug Subst. 1981, 9, 295–340. [Google Scholar] [CrossRef]
- Jao, R.L.; Jackson, G.G. Gentamicin Sulfate, New Antibiotic against Gram-Negative Bacilli. JAMA 1964, 189, 817–822. [Google Scholar] [CrossRef]
- Weinstein, M.J.; Luedemann, G.M.; Oden, E.M.; Wagman, G.H.; Rosselet, J.P.; Marquez, J.A.; Coniglio, C.T.; Charney, W.; Herzog, H.L.; Black, J. Gentamicin,1a New Antibiotic Complex from Micromonospora. J. Med. Chem. 1963, 6, 463–464. [Google Scholar] [CrossRef]
- Pourfeizi, H.H.; Saleh, P.; Abbasalizadeh, S.; Rezaeian, S.; Naghavi-Behzad, M.; Piri, R. Gentamicin-mediated ototoxicity and nephrotoxicity: A clinical trial study. Niger. Med. J. 2016, 57, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Weng, X.; Neethirajan, S. Ensuring food safety: Quality monitoring using microfluidics. Trends Food Sci. Technol. 2017, 65, 10–22. [Google Scholar] [CrossRef]
- De Oliveira, R.A.; Camargo, F.; Pesquero, N.C.; Faria, R.C. A simple method to produce 2D and 3D microfluidic paper-based analytical devices for clinical analysis. Anal. Chim. Acta 2017, 957, 40–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, C.K. Aptasensors—The future of biosensing? Anal. Bioanal. Chem. 2002, 372, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Klug, S.J.; Famulok, M. All you wanted to know about SELEX. Mol. Biol. Rep. 1994, 20, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, B.; Angelomé, P.C.; Lechuga, L.M.; Liz-Marzán, L.M. LSPR-based nanobiosensors. Nano Today 2009, 4, 244–251. [Google Scholar] [CrossRef]
- Hu, X.; Chang, K.; Wang, S.; Sun, X.; Hu, J.; Jiang, M. Aptamer-functionalized AuNPs for the high-sensitivity colorimetric detection of melamine in milk samples. PLoS ONE 2018, 13, e0201626. [Google Scholar] [CrossRef] [Green Version]
- Song, K.-M.; Cho, M.; Ban, C.; Min, K.; Jeon, S.H.; Kim, T.; Han, M.S.; Ku, J.K.; Ban, C. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer. Anal. Biochem. 2011, 415, 175–181. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, Y.; Jia, J.; Xiang, Y. Colorimetric aptasensors for determination of tobramycin in milk and chicken eggs based on DNA and gold nanoparticles. Food Chem. 2018, 249, 98–103. [Google Scholar] [CrossRef]
- Rowe, A.A.; Miller, E.A.; Plaxco, K.W. Reagentless Measurement of Aminoglycoside Antibiotics in Blood Serum via an Electrochemical, Ribonucleic Acid Aptamer-Based Biosensor. Anal. Chem. 2010, 82, 7090–7095. [Google Scholar] [CrossRef] [Green Version]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of Size and Concentration of Gold Nanoparticles from UV−Vis Spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Weng, X.; Ahmed, S.R.; Neethirajan, S. A nanocomposite-based biosensor for bovine haptoglobin on a 3D paper-based analytical device. Sens. Actuators B Chem. 2018, 265, 242–248. [Google Scholar] [CrossRef]
- Emrani, A.S.; Danesh, N.M.; Lavaee, P.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Colorimetric and fluorescence quenching aptasensors for detection of streptomycin in blood serum and milk based on double-stranded DNA and gold nanoparticles. Food Chem. 2016, 190, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Musile, G.; Mccord, B. An aptamer-based paper microfluidic device for the colorimetric determination of cocaine. Electrophoresis 2017, 39, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Soda, Y.; Bakker, E. Quantification of Colorimetric Data for Paper-Based Analytical Devices. ACS Sens. 2019, 4, 3093–3101. [Google Scholar] [CrossRef]
- Zeng, S.; Yong, K.-T.; Roy, I.; Dinh, X.-Q.; Yu, X.; Luan, F. A Review on Functionalized Gold Nanoparticles for Biosensing Applications. Plasmonics 2011, 6, 491–506. [Google Scholar] [CrossRef]
- Cao, J.; Sun, T.; Grattan, K.T. Gold nanorod-based localized surface plasmon resonance biosensors: A review. Sens. Actuators B Chem. 2014, 195, 332–351. [Google Scholar] [CrossRef]
- Pandana, H.; Aschenbach, K.H.; Gomez, R.D. Systematic Aptamer-Gold Nanoparticle Colorimetry for Protein Detection: Thrombin. IEEE Sens. J. 2008, 8, 661–666. [Google Scholar] [CrossRef]
- Ha, N.-R.; Jung, I.-P.; Kim, S.-H.; Kim, A.-R.; Yoon, M.-Y. Paper chip-based colorimetric sensing assay for ultra-sensitive detection of residual kanamycin. Process. Biochem. 2017, 62, 161–168. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramalingam, S.; Collier, C.M.; Singh, A. A Paper-Based Colorimetric Aptasensor for the Detection of Gentamicin. Biosensors 2021, 11, 29. https://0-doi-org.brum.beds.ac.uk/10.3390/bios11020029
Ramalingam S, Collier CM, Singh A. A Paper-Based Colorimetric Aptasensor for the Detection of Gentamicin. Biosensors. 2021; 11(2):29. https://0-doi-org.brum.beds.ac.uk/10.3390/bios11020029
Chicago/Turabian StyleRamalingam, Saipriya, Christopher M. Collier, and Ashutosh Singh. 2021. "A Paper-Based Colorimetric Aptasensor for the Detection of Gentamicin" Biosensors 11, no. 2: 29. https://0-doi-org.brum.beds.ac.uk/10.3390/bios11020029