Machine Learning-Based Identification of Colon Cancer Candidate Diagnostics Genes
Abstract
:Simple Summary
Abstract
1. Introduction
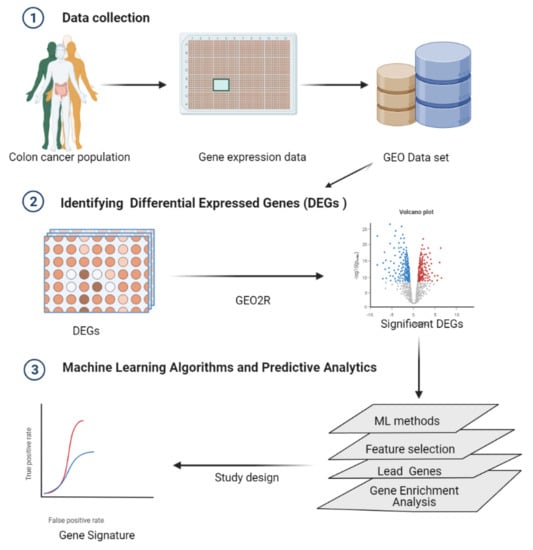
2. Methods
2.1. Data
Differentially Expressed Genes (DEGs) Identified by GEO2R
2.2. Machine Learning Algorithms and Predictive Analytics
2.2.1. Hyperparameter Optimization
2.2.2. Machine Learning Model Evaluation
2.2.3. Feature Selection
2.3. Gene Enrichment Analysis
2.3.1. Association of the Gene Markers with miRNA
2.3.2. Sample Size Estimates for Future Validation Experiments
3. Results
3.1. Differential Expressed Genes (DEGs)
Performance Evaluation
3.2. Gene Selection
Gene Ontology (GO) Enrichment Analysis
3.3. Associating Selected Genes with miRNA Using NetworkAnalyst
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUROC | Area under the receiver operating characteristic curve |
| CRC | Colon cancer |
| DEGs | Differential expressed genes |
| GEO | Gene Expression Omnibus |
| GO | Gene ontology |
| miRNA | microRNA |
| MDI | Mean decrease in impurity |
| RF | Random forest |
References
- Siegel, R.; DeSantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA A Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Worldwide incidence and mortality of colorectal cancer and human development index (HDI): An ecological study. WCRJ 2019, 6, 1433.
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, J.; Prenen, H. Molecular genetics of colorectal cancer. Ann. Gastroenterol. 2014, 27, 9–14. [Google Scholar]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Stefano, G.B.; Mantione, K.J.; Kream, R.M.; Kuzelova, H.; Ptacek, R.; Raboch, J.; Samuel, J.M. Comparing Bioinformatic Gene Expression Profiling Methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [Google Scholar] [CrossRef] [Green Version]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-Y.; Lee, S.-G.; Oh, T.-J.; Lim, S.R.; Kim, S.-H.; Lee, H.J.; Kim, Y.-S.; Choi, H.-K. Antiproliferative and Apoptotic Activity of Chamaecyparis obtusa Leaf Extract against the HCT116 Human Colorectal Cancer Cell Line and Investigation of the Bioactive Compound by Gas Chromatography-Mass Spectrometry-Based Metabolomics. Molecules 2015, 20, 18066–18082. [Google Scholar] [CrossRef] [Green Version]
- Dalal, N.; Jalandra, R.; Sharma, M.; Prakash, H.; Makharia, G.K.; Solanki, P.R.; Singh, R.; Kumar, A. Omics technologies for improved diagnosis and treatment of colorectal cancer: Technical advancement and major perspectives. Biomed. Pharmacother. 2020, 131, 110648. [Google Scholar] [CrossRef]
- Chen, M.; Yang, X.; Yang, M.; Zhang, W.; Li, L.; Sun, Q. Identification of a novel biomarker-CCL5 using antibody microarray for colorectal cancer. Pathol. Res. Pract. 2019, 215, 1033–1037. [Google Scholar] [CrossRef]
- Wei, F.-Z.; Mei, S.-W.; Wang, Z.-J.; Chen, J.-N.; Shen, H.-Y.; Zhao, F.-Q.; Li, J.; Liu, Z.; Liu, Q. Differential Expression Analysis Revealing CLCA1 to Be a Prognostic and Diagnostic Biomarker for Colorectal Cancer. Front. Oncol. 2020, 10, 573295. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Wang, X.; Yang, Q. CDK1 and CDC20 overexpression in patients with colorectal cancer are associated with poor prognosis: Evidence from integrated bioinformatics analysis. World J. Surg. Oncol. 2020, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pons, M.; Cruz-Correa, M. Colorectal Cancer Biomarkers: Where Are We Now? BioMed. Res. Int. 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.-R.; Huang, M.-Y.; Chang, H.-J. Molecular Detection of Circulating Tumor Cells With Multiple mRNA Markers by Genechip for Colorectal Cancer Early Diagnosis and Prognosis Prediction. Genom. Med. Biomark. Health Sci. 2011, 3, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Dasí, F.; Lledó, S.; García-Granero, E.; Ripoll, R.; Marugán, M.; Tormo, M.; García-Conde, J.; Aliño, S.F. Real-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: A simple blood test to monitor disease in cancer patients. Lab. Investig. 2001, 81, 767–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiedeck, T.H.K.; Wellm, C.; Roblick, U.J.; Broll, R.; Bruch, H.-P. Diagnosis and Monitoring of Colorectal Cancer by L6 Blood Serum Polymerase Chain Reaction Is Superior to Carcinoembryonic Antigen-Enzyme-Linked Immunosorbent Assay. Dis. Colon Rectum 2003, 46, 818–825. [Google Scholar] [CrossRef]
- Liu, X.; Bing, Z.; Wu, J.; Zhang, J.; Zhou, W.; Ni, M.; Meng, Z.; Liu, S.; Tian, J.; Zhang, X.; et al. Integrative Gene Expression Profiling Analysis to Investigate Potential Prognostic Biomarkers for Colorectal Cancer. Med. Sci. Monit. 2020, 26, e918906. [Google Scholar] [CrossRef]
- Torres, S.; Bartolome, R.A.; Mendes, M.; Barderas, R.; Fernández-Aceñerp, M.J.; Peláez-García, A.; Peña, C.; Lopez-Lucendo, M.; Villar-Vázquez, R.; De Herreros, A.G.; et al. Proteome Profiling of Cancer-Associated Fibroblasts Identifies Novel Proinflammatory Signatures and Prognostic Markers for Colorectal Cancer. Clin. Cancer Res. 2013, 19, 6006–6019. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.R.; Kwon, H.N.; Nam, H.; Kim, J.J.; Park, S.; Kim, Y.-H. Urine-NMR metabolomics for screening of advanced colorectal adenoma and early stage colorectal cancer. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Schirripa, M.; Lenz, H.-J. Biomarker in Colorectal Cancer. Cancer J. 2016, 22, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.; Lin, W.; Zhao, X.-M. Identifying Molecular Biomarkers for Diseases With Machine Learning Based on Integrative Omics. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, M.-J.; Ping, J. Clinicopathological Features and Survival Outcomes of Colorectal Cancer in Young Versus Elderly: A Population-Based Cohort Study of SEER 9 Registries Data (1988–2011). Medicine 2015, 94, e1402. [Google Scholar] [CrossRef] [PubMed]
- Mangone, L.; Pinto, C.; Mancuso, P.; Ottone, M.; Bisceglia, I.; Chiaranda, G.; Michiara, M.; Vicentini, M.; Carrozzi, G.; Ferretti, S.; et al. Colon cancer survival differs from right side to left side and lymph node harvest number matter. BMC Public Health 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, B.M.; Zanetti, K.A.; Robles, A.; Schetter, A.J.; Goodman, J.; Hayes, R.; Huang, W.-Y.; Gunter, M.J.; Yeager, M.; Burdette, L.; et al. Germline variation inNCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. Int. J. Cancer 2014, 134, 1399–1407. [Google Scholar] [CrossRef] [Green Version]
- Skrzypczak, M.; Goryca, K.; Rubel, T.; Paziewska, A.; Mikula, M.; Jarosz, D.; Pachlewski, J.; Oledzki, J.; Ostrowsk, J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE 2010, 5, e13091. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D9955. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Additive logistic regression: A statistical view of boosting. Ann. Stat. 2000, 28, 337–407. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Yuan, Z.; Ghosh, D. Combining Multiple Biomarker Models in Logistic Regression. Biometrics 2008, 64, 431–439. [Google Scholar] [CrossRef]
- Tolles, J.; Meurer, W.J. Logistic Regression: Relating Patient Characteristics to Outcomes. JAMA 2016, 316, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Sambo, F.; Trifoglio, E.; Di Camillo, B.; Toffolo, G.M.; Cobelli, C. Bag of Naïve Bayes: Biomarker selection and classification from genome-wide SNP data. BMC Bioinform. 2012, 13, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ishwaran, H. Random forests for genomic data analysis. Genomics 2012, 99, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Yin, Y.; Quan, X.; Zhang, H. Gene Expression Value Prediction Based on XGBoost Algorithm. Front. Genet. 2019, 10, 1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Bauer, E.; Kohavi, R. An Empirical Comparison of Voting Classification Algorithms: Bagging, Boosting, and Variants. Mach. Learn. 1999, 36, 105–139. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Geurts, P.; Maree, R.; Wehenkel, L. Extremely Randomized Trees and Random Subwindows for Image Classification, Annotation, and Retrieval. Mach. Learn. 2013, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Schapire, R.E. Explaining AdaBoost. In Empirical Inference; Springer: Berlin/Heidelberg, Germany, 2013; pp. 37–52. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; KDD ’16, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Davis, J.; Goadrich, M. The Relationship Between Precision-Recall and ROC Curves. In Proceedings of the 23rd International Conference on Machine Learning; Association for Computing Machinery: New York, NY, USA, 2006; pp. 233–240. [Google Scholar]
- Hand, D.J. Assessing the Performance of Classification Methods. Int. Stat. Rev. 2012, 80, 400–414. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In AI 2006: Advances in Artificial Intelligence; Lecture Notes in Computer Science; Sattar, A., Kang, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4304, pp. 1015–1021. ISBN 978-3-540-49787-5. [Google Scholar]
- Gilles, L.; Wehenkel, L.; Sutera, A.; Geurts, P. Understanding variable importances in forests of randomized trees. In Proceedings of the Twenty-Seventh Conference on Neural Information Processing Systems—NIPS, Lake Tahoe, CA, USA, 5–10 December 2013. [Google Scholar]
- Kursa, M.B.; Jankowski, A.; Rudnicki, W.R. Boruta—A System for Feature Selection. Fundam. Inform. 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Sandri, M.; Zuccolotto, P. A Bias Correction Algorithm for the Gini Variable Importance Measure in Classification Trees. J. Comput. Graph. Stat. 2008, 17, 611–628. [Google Scholar] [CrossRef]
- Chen, R.-C.; Dewi, C.; Huang, S.-W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. J. Big Data 2020, 7, 1–26. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- Acharjee, A.; Larkman, J.; Xu, Y.; Cardoso, V.R.; Gkoutos, G.V. A random forest based biomarker discovery and power analysis framework for diagnostics research. BMC Med. Genom. 2020, 13, 1–14. [Google Scholar] [CrossRef]
- Shafiha, R.; Bahcivanci, B.; Gkoutos, G.V.; Acharjee, A. Machine Learning-Based Identification of Potentially Novel Non-Alcoholic Fatty Liver Disease Biomarkers. Biomedicines 2021, 9, 1636. [Google Scholar] [CrossRef]
- Acharjee, A.; Ament, Z.; West, J.A.; Stanley, E.; Griffin, J.L. Integration of metabolomics, lipidomics and clinical data using a machine learning method. BMC Bioinform. 2016, 17 (Suppl. S15), 440. [Google Scholar] [CrossRef] [Green Version]
- Quraishi, M.N.; Acharjee, A.; Beggs, A.D.; Horniblow, R.; Tselepis, C.; Gkoutos, G.; Ghosh, S.; Rossiter, A.E.; Loman, N.; van Schaik, W.; et al. A Pilot Integrative Analysis of Colonic Gene Expression, Gut Microbiota, and Immune Infiltration in Primary Sclerosing Cholangitis-Inflammatory Bowel Disease: Association of Disease With Bile Acid Pathways. J. Crohn’s Colitis 2020, 14, 935–947. [Google Scholar] [CrossRef] [Green Version]
- Frank, H. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis, 2nd ed.; Springer: New York, NY, USA, 2015. [Google Scholar]
- Kumara, H.S.; Bellini, G.A.; Caballero, O.L.; Herath, S.A.; Su, T.; Ahmed, A.; Njoh, L.; Cekic, V.; Whelan, R.L. P-Cadherin (CDH3) is overexpressed in colorectal tumors and has potential as a serum marker for colorectal cancer monitoring. Oncoscience 2017, 4, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhao, J.; Dai, X.; Xie, Y.; Dong, M. High expression of CDH3 predicts a good prognosis for colon adenocarcinoma patients. Exp. Ther. Med. 2019, 18, 841–847. [Google Scholar] [CrossRef]
- Hahn-Strömberg, V.; Askari, S.; Ahmad, A.; Befekadu, R.; Nilsson, T.K. Expression of claudin 1, claudin 4, and claudin 7 in colorectal cancer and its relation with CLDN DNA methylation patterns. Tumor Biol. 2017, 39, 1010428317697569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Liu, Q.-M.; Du, P.-C.; Ning, D.; Mo, J.; Zhu, H.-D.; Wang, C.; Ge, Q.-Y.; Cheng, Q.; Zhang, X.-W.; et al. Type-2 11β-hydroxysteroid dehydrogenase promotes the metastasis of colorectal cancer via the Fgfbp1-AKT pathway. Am. J. Cancer Res. 2020, 10, 662–673. [Google Scholar] [PubMed]
- Yang, G.-Z.; Hu, L.; Cai, J.; Chen, H.-Y.; Zhang, Y.; Feng, D.; Qi, C.-Y.; Zhai, Y.-X.; Gong, H.; Fu, H.; et al. Prognostic value of carbonic anhydrase VII expression in colorectal carcinoma. BMC Cancer 2015, 15, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Chen, H.-Y.; Han, T.; Yang, G.-Z.; Feng, D.; Qi, C.-Y.; Gong, H.; Zhai, Y.-X.; Cai, Q.-P.; Gao, C.-F. Downregulation of DHRS9 expression in colorectal cancer tissues and its prognostic significance. Tumor Biol. 2015, 37, 837–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, S.; Huang, Y.; Shi, M.; Qian, X.; Li, H.; Peng, C.; Kong, B.; Zou, X.; Shen, S. Protective role of ABCG2 against oxidative stress in colorectal cancer and its potential underlying mechanism. Oncol. Rep. 2018, 40, 2137–2146. [Google Scholar] [CrossRef] [PubMed]
- Expression of ABCG2 and its Significance in Colorectal Cancer. Asian Pac. J. Cancer Prev. 2010, 11, 845–848.
- Tuy, H.D.; Shiomi, H.; Mukaisho, K.I.; Naka, S.; Shimizu, T.; Sonoda, H.; Mekata, E.; Endo, Y.; Kurumi, Y.; Sugihara, H.; et al. ABCG2 expression in colorectal adenocarcinomas may predict resistance to irinotecan. Oncol. Lett. 2016, 12, 2752–2760. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Ma, J.; Zhou, W.; Li, Z.; Zhou, X.; Cao, B.; Zhang, Y.; Liu, J.; Yang, Z.; Zhang, H.; et al. Identification of hub genes and outcome in colon cancer based on bioinformatics analysis. Cancer Manag. Res. 2018, 11, 323–338. [Google Scholar] [CrossRef] [Green Version]
- Pira, G.; Uva, P.; Scanu, A.M.; Rocca, P.C.; Murgia, L.; Uleri, E.; Piu, C.; Porcu, A.; Carru, C.; Manca, A.; et al. Landscape of transcriptome variations uncovering known and novel driver events in colorectal carcinoma. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Coller, H.A. Is Cancer a Metabolic Disease? Am. J. Pathol. 2014, 184, 4–17. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Rokavec, M.; Hermeking, H. Soluble IL6R represents a miR-34a target: Potential implications for the recently identified IL-6R/STAT3/miR-34a feed-back loop. Oncotarget 2015, 6, 14026–14032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vainer, N.; Dehlendorff, C.; Johansen, J.S. Systematic literature review of IL-6 as a biomarker or treatment target in patients with gastric, bile duct, pancreatic and colorectal cancer. Oncotarget 2018, 9, 29820–29841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, G.; Wang, L.; Wen, Y.; Ren, X.; Zuo, S. Identification of key genes for predicting colorectal cancer prognosis by integrated bioinformatics analysis. Oncol. Lett. 2019, 19, 388–398. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Liu, H.; Lin, H.-C.; Gan, D.; Jin, W.; Cui, C.; Yan, Y.; Qian, Y.; Han, C.; Wang, Z. Association of a novel seven-gene expression signature with the disease prognosis in colon cancer patients. Aging 2019, 11, 8710–8727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tan, P.; Zhuang, Y.; Du, L. hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am. J. Physiol. Liver Physiol. 2020, 319, G703–G717. [Google Scholar] [CrossRef] [PubMed]
- Mencia, N.; Selga, E.; Noe, V.; Ciudad, C.J. Underexpression of miR-224 in methotrexate resistant human colon cancer cells. Biochem. Pharmacol. 2011, 82, 1572–1582. [Google Scholar] [CrossRef]
- Andersen, V.; Vogel, L.K.; Kopp, T.I.; Sæbø, M.; Nonboe, A.W.; Hamfjord, J.; Kure, E.H.; Vogel, U. High ABCC2 and Low ABCG2 Gene Expression Are Early Events in the Colorectal Adenoma-Carcinoma Sequence. PLoS ONE 2015, 10, e0119255. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S. Upregulated miR-132 in Lgr5+gastric cancer stem cell-like cells contributes to cisplatin-resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Cherradi, S.; Ayrolles-Torro, A.; Vezzo-Vié, N.; Gueguinou, N.; Denis, V.; Combes, E.; Boissière, F.; Busson, M.; Canterel-Thouennon, L.; Mollevi, C.; et al. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J. Exp. Clin. Cancer Res. 2017, 36, 89. [Google Scholar] [CrossRef] [Green Version]
- Miwa, N.; Furuse, M.; Tsukita, S.; Niikawa, N.; Nakamura, Y.; Furukawa, Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol. Res. 2001, 12, 469–476. [Google Scholar] [CrossRef]
- Singh, A.B.; Sharma, A.; Smith, J.J.; Krishnan, M.; Chen, X.; Eschrich, S.; Washington, M.K.; Yeatman, T.J.; Beauchamp, R.D.; Dhawan, P. Claudin-1 Up-regulates the Repressor ZEB-1 to Inhibit E-Cadherin Expression in Colon Cancer Cells. Gastroenterology 2011, 141, 2140–2153. [Google Scholar] [CrossRef] [Green Version]
- Bravo-Merodio, L.; Acharjee, A.; Russ, D.; Bisht, V.; Williams, J.A.; Tsaprouni, L.G.; Gkoutos, G.V. Translational biomarkers in the era of precision medicine. Int. Rev. Cytol. 2021, 102, 191–232. [Google Scholar] [CrossRef]
- Bailey, J.R.; Aggarwal, A.; Imperiale, T.F. Colorectal Cancer Screening: Stool DNA and Other Noninvasive Modalities. Gut Liver 2016, 10, 204–211. [Google Scholar] [CrossRef] [PubMed]
- de Wit, M.; Fijneman, R.J.; Verheul, H.M.; Meijer, G.A.; Jimenez, C.R. Proteomics in colorectal cancer translational research: Biomarker discovery for clinical applications. Clin. Biochem. 2013, 46, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Chaver, P.; Otero-Estévez, O.; Páez de la Cadena, M.; Rodríguez-Berrocal, F.J.; Martínez-Zorzano, V.S. Proteomics for discovery of candidate colorectal cancer biomarkers. World J. Gastroenterol. 2014, 20, 3804–3824. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koppad, S.; Basava, A.; Nash, K.; Gkoutos, G.V.; Acharjee, A. Machine Learning-Based Identification of Colon Cancer Candidate Diagnostics Genes. Biology 2022, 11, 365. https://0-doi-org.brum.beds.ac.uk/10.3390/biology11030365
Koppad S, Basava A, Nash K, Gkoutos GV, Acharjee A. Machine Learning-Based Identification of Colon Cancer Candidate Diagnostics Genes. Biology. 2022; 11(3):365. https://0-doi-org.brum.beds.ac.uk/10.3390/biology11030365
Chicago/Turabian StyleKoppad, Saraswati, Annappa Basava, Katrina Nash, Georgios V. Gkoutos, and Animesh Acharjee. 2022. "Machine Learning-Based Identification of Colon Cancer Candidate Diagnostics Genes" Biology 11, no. 3: 365. https://0-doi-org.brum.beds.ac.uk/10.3390/biology11030365