Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine
Abstract
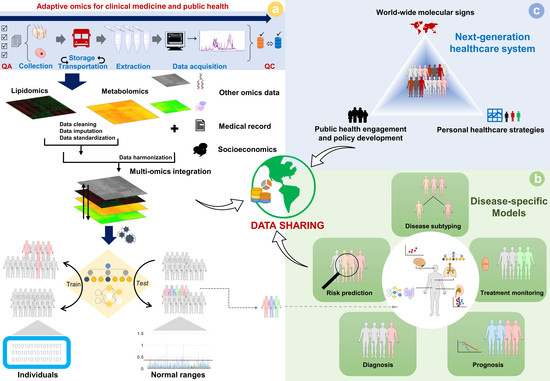
:1. Introduction
2. Representative Achievements of Metabolomics in Public Health and Clinical Research
3. Analytical and Computational Challenges in Clinical Metabolomics and Lipidomics Studies
3.1. The Need for Standardized Biobank Establishment
3.2. The High Demand for Quality Assurance and Quality Control Standards
3.3. The Role of Establishing an Automated System for Sample Handling, Data Acquisition, and Automated Modeling
3.4. The Assessment of Systematic Errors in Clinical Metabolomics Studies
3.5. The Advancement and Limitations of Metabolite Annotation/Identification Standards
3.6. Ongoing Efforts for a Universal Workflow Covering all Computational Steps of Metabolomics Research
4. Specific Notes on Clinical Lipidomics Studies
4.1. QA/QC and Batch Effect Removal for Large-Scale Lipidomics Studies
4.2. The Need for High-Confidence Lipid Identification and Quantification
5. Toward Reproducible Data Analysis and Interpretation in Clinical Metabolomics and Lipidomics
6. Fundamental Requirements on Standards for Reporting Metabolomics-Based Biomarker Studies at Clinical and Epidemiological Scale
7. Multi-Omics Integration for Precision Medicine
8. Potential Bioethical Issues Associated with the Sharing of Clinical Metabolomics and Lipidomics Data
9. Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bizzarri, M.; Brash, D.E.; Briscoe, J.; Grieneisen, V.A.; Stern, C.D.; Levin, M. A call for a better understanding of causation in cell biology. Nat. Rev. Mol. Cell Biol. 2019, 20, 261–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Snyder, M. Promise of personalized omics to precision medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013, 5, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. OMICS 2018, 22, 630–636. [Google Scholar] [CrossRef] [Green Version]
- Sampson, J.N.; Boca, S.M.; Shu, X.O.; Stolzenberg-Solomon, R.Z.; Matthews, C.E.; Hsing, A.W.; Tan, Y.T.; Ji, B.-T.; Chow, W.-H.; Cai, Q.; et al. Metabolomics in epidemiology: Sources of variability in metabolite measurements and implications. Cancer Epidemiol. Biomark. Prev. 2013, 22, 631. [Google Scholar] [CrossRef] [Green Version]
- Ala-Korpela, M.; Davey Smith, G. Metabolic profiling–multitude of technologies with great research potential, but (when) will translation emerge? Int. J. Epidemiol. 2016, 45, 1311–1318. [Google Scholar] [CrossRef] [Green Version]
- Valdes, A.; Tzoulaki, I.; Ioannidis, J.P.A.; Elliott, P.; Ebbels, T.M.D. Design and analysis of metabolomics studies in epidemiologic research: A primer on -omic technologies. Am. J. Epidemiol. 2014, 180, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Fearnley, L.G.; Inouye, M. Metabolomics in epidemiology: From metabolite concentrations to integrative reaction networks. Int. J. Epidemiol. 2016, 45, 1319–1328. [Google Scholar] [CrossRef] [Green Version]
- Mundra, P.A.; Shaw, J.E.; Meikle, P.J. Lipidomic analyses in epidemiology. Int. J. Epidemiol. 2016, 45, 1329–1338. [Google Scholar] [CrossRef] [Green Version]
- van Roekel, E.H.; Loftfield, E.; Kelly, R.S.; Zeleznik, O.A.; Zanetti, K.A. Metabolomics in epidemiologic research: Challenges and opportunities for early-career epidemiologists. Metabolomics 2019, 15, 9. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, C.; Hernández-Alonso, P.; Bulló, M.; Ruiz-Canela, M.; Yu, E.; Guasch-Ferré, M.; Toledo, E.; Dennis, C.; Deik, A.; Clish, C.; et al. Plasma metabolites associated with coffee consumption: A metabolomic approach within the predimed study. Nutrients 2019, 11, 1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesch, B.; Brüning, T.; Johnen, G.; Casjens, S.; Bonberg, N.; Taeger, D.; Müller, A.; Weber, D.G.; Behrens, T. Biomarker research with prospective study designs for the early detection of cancer. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Fest, J.; Vijfhuizen, L.S.; Goeman, J.J.; Veth, O.; Joensuu, A.; Perola, M.; Mannisto, S.; Ness-Jensen, E.; Hveem, K.; Haller, T.; et al. Search for early pancreatic cancer blood biomarkers in five European prospective population biobanks using metabolomics. Endocrinology 2019, 160, 1731–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, R.A.; Pan, K.; Harville, E.W.; McRitchie, S.; Sumner, S. Metabolomics to reveal biomarkers and pathways of preterm birth: A systematic review and epidemiologic perspective. Metabolomics 2019, 15, 124. [Google Scholar] [CrossRef] [PubMed]
- Playdon, M.C.; Joshi, A.D.; Tabung, F.K.; Cheng, S.; Henglin, M.; Kim, A.; Lin, T.; van Roekel, E.H.; Huang, J.; Krumsiek, J.; et al. Metabolomics analytics workflow for epidemiological research: Perspectives from the Consortium of Metabolomics Studies (COMETS). Metabolites 2019, 9, 145. [Google Scholar] [CrossRef] [Green Version]
- Chu, S.H.; Huang, M.; Kelly, R.S.; Benedetti, E.; Siddiqui, J.K.; Zeleznik, O.A.; Pereira, A.; Herrington, D.; Wheelock, C.E.; Krumsiek, J.; et al. Integration of metabolomic and other omics data in population-based study designs: An epidemiological perspective. Metabolites 2019, 9, 117. [Google Scholar] [CrossRef] [Green Version]
- Cuperlovic-Culf, M. Machine learning methods for analysis of metabolic data and metabolic pathway modeling. Metabolites 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Ciocan-Cartita, A.C.; Jurj, A.; Buse, M.; Gulei, D.; Braicu, C.; Raduly, L.; Cojocneanu, R.; Pruteanu, L.L.; Iuga, A.C.; Coza, O.; et al. The relevance of mass spectrometry analysis for personalized medicine through its successful application in cancer “omics”. Int. J. Mol. Sci. 2019, 20, 2576. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Shen, J.; Moore, S.C.; Ye, Y.; Wu, X.; Esteva, F.J.; Tripathy, D.; Chow, W.H. Breast cancer risk in relation to plasma metabolites among Hispanic and African American women. Breast Cancer Res. Treat. 2019, 176, 687–696. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, H.; Zhang, X.; Zhao, X.; Song, Z.; Ouyang, J. Metabolic discrimination of breast cancer subtypes at the single-cell level by multiple microextraction coupled with mass spectrometry. Anal. Chem. 2019, 91, 3667–3674. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.T.; Showalter, M.R.; Fiehn, O. Inborn errors of metabolism in the era of untargeted metabolomics and lipidomics. Metabolites 2019, 9, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anh, N.H.; Long, N.P.; Kim, S.J.; Min, J.E.; Yoon, S.J.; Kim, H.M.; Yang, E.; Hwang, E.S.; Park, J.H.; Hong, S.S.; et al. Steroidomics for the prevention, assessment, and management of cancers: A systematic review and functional analysis. Metabolites 2019, 9, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.J.; Cho, E.J.; Yu, K.S.; Jang, I.J.; Yoon, J.H.; Park, T.; Cho, J.Y. Comprehensive metabolomic search for biomarkers to differentiate early stage hepatocellular carcinoma from cirrhosis. Cancers 2019, 11, 1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guasch-Ferre, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Hu, F.B. Metabolomics in prediabetes and diabetes: A systematic review and meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef] [Green Version]
- Arneth, B.; Arneth, R.; Shams, M. Metabolomics of type 1 and type 2 diabetes. Int. J. Mol. Sci. 2019, 20, 2467. [Google Scholar] [CrossRef] [Green Version]
- Arlt, W.; Biehl, M.; Taylor, A.E.; Hahner, S.; Libe, R.; Hughes, B.A.; Schneider, P.; Smith, D.J.; Stiekema, H.; Krone, N.; et al. Urine steroid metabolomics as a biomarker tool for detecting malignancy in adrenal tumors. J. Clin. Endocrinol. Metab. 2011, 96, 3775–3784. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448. [Google Scholar] [CrossRef]
- Kerkhofs, T.M.A.; Kerstens, M.N.; Kema, I.P.; Willems, T.P.; Haak, H.R. Diagnostic value of urinary steroid profiling in the evaluation of adrenal tumors. Horm. Cancer 2015, 6, 168–175. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Lin, W.; Broadhurst, D.; Begley, P.; Brown, M.; Zelena, E.; Vaughan, A.A.; Halsall, A.; Harding, N.; Knowles, J.D.; et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics 2015, 11, 9–26. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, R.G.; Fuhrman, B.J.; Moore, S.C.; Matthews, C.E. Epidemiologic studies of estrogen metabolism and breast cancer. Steroids 2015, 99, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rewers, M.; Hyoty, H.; Lernmark, A.; Hagopian, W.; She, J.X.; Schatz, D.; Ziegler, A.G.; Toppari, J.; Akolkar, B.; Krischer, J. The Environmental Determinants of Diabetes in the Young (TEDDY) study: 2018 Update. Curr. Diabetes Rep. 2018, 18, 136. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; Fernández-Real, J.-M.; Federici, M.; Serino, M.; Abbott, J.; Charpentier, J.; Heymes, C.; Luque, J.L.; Anthony, E.; Barton, R.H.; et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018, 24, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Hirayama, A.; Chan, Q.; Kurihara, A.; Fukai, K.; Iida, M.; Kato, S.; Sugiyama, D.; Kuwabara, K.; Takeuchi, A.; et al. Reliability of plasma polar metabolite concentrations in a large-scale cohort study using capillary electrophoresis-mass spectrometry. PLoS ONE 2018, 13, e0191230. [Google Scholar] [CrossRef] [PubMed]
- Deelen, J.; Kettunen, J.; Fischer, K.; van der Spek, A.; Trompet, S.; Kastenmuller, G.; Boyd, A.; Zierer, J.; van den Akker, E.B.; Ala-Korpela, M.; et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 2019, 10, 3346. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, D.K.; Sinclair, E.; Xu, Y.; Sarkar, D.; Walton-Doyle, C.; Liscio, C.; Banks, P.; Milne, J.; Silverdale, M.; Kunath, T.; et al. Discovery of volatile biomarkers of Parkinson’s disease from sebum. ACS Cent. Sci. 2019, 5, 599–606. [Google Scholar] [CrossRef] [Green Version]
- Tzoulaki, I.; Castagné, R.; Boulangé, C.L.; Karaman, I.; Chekmeneva, E.; Evangelou, E.; Ebbels, T.M.D.; Kaluarachchi, M.R.; Chadeau-Hyam, M.; Mosen, D.; et al. Serum metabolic signatures of coronary and carotid atherosclerosis and subsequent cardiovascular disease. Eur. Heart J. 2019, 40, 2883–2896. [Google Scholar] [CrossRef] [Green Version]
- Cirulli, E.T.; Guo, L.; Leon Swisher, C.; Shah, N.; Huang, L.; Napier, L.A.; Kirkness, E.F.; Spector, T.D.; Caskey, C.T.; Thorens, B.; et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019, 29, 488–500. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.Z.; Liu, J.; Morningstar, J.; Heckman-Stoddard, B.M.; Lee, C.G.; Dagogo-Jack, S.; Ferguson, J.F.; Hamman, R.F.; Knowler, W.C.; Mather, K.J.; et al. Metabolite Profiles of Incident Diabetes and Heterogeneity of Treatment Effect in the Diabetes Prevention Program. Diabetes 2019, 68, 2337–2349. [Google Scholar] [CrossRef]
- Gomez, C.; Gonzalez-Riano, C.; Barbas, C.; Kolmert, J.; Hyung Ryu, M.; Carlsten, C.; Dahlen, S.E.; Wheelock, C.E. Quantitative metabolic profiling of urinary eicosanoids for clinical phenotyping. J. Lipid Res. 2019, 60, 1164–1173. [Google Scholar] [CrossRef]
- Yu, Z.; Zhai, G.; Singmann, P.; He, Y.; Xu, T.; Prehn, C.; Romisch-Margl, W.; Lattka, E.; Gieger, C.; Soranzo, N.; et al. Human serum metabolic profiles are age dependent. Aging Cell 2012, 11, 960–967. [Google Scholar] [CrossRef]
- Meikle, P.J.; Wong, G.; Barlow, C.K.; Weir, J.M.; Greeve, M.A.; MacIntosh, G.L.; Almasy, L.; Comuzzie, A.G.; Mahaney, M.C.; Kowalczyk, A.; et al. Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS ONE 2013, 8, e74341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganna, A.; Salihovic, S.; Sundstrom, J.; Broeckling, C.D.; Hedman, A.K.; Magnusson, P.K.; Pedersen, N.L.; Larsson, A.; Siegbahn, A.; Zilmer, M.; et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014, 10, e1004801. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, C.; Pechlaner, R.; Willeit, P.; Langley, S.R.; Mangino, M.; Mayr, U.; Menni, C.; Moayyeri, A.; Santer, P.; Rungger, G.; et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014, 129, 1821–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loftfield, E.; Rothwell, J.A.; Sinha, R.; Keski-Rahkonen, P.; Robinot, N.; Albanes, D.; Weinstein, S.J.; Derkach, A.; Sampson, J.; Scalbert, A.; et al. Prospective investigation of serum metabolites, coffee drinking, liver cancer incidence, and liver disease mortality. J. Natl. Cancer Inst. 2019, 112. [Google Scholar] [CrossRef]
- Floegel, A.; Kuhn, T.; Sookthai, D.; Johnson, T.; Prehn, C.; Rolle-Kampczyk, U.; Otto, W.; Weikert, C.; Illig, T.; von Bergen, M.; et al. Serum metabolites and risk of myocardial infarction and ischemic stroke: A targeted metabolomic approach in two German prospective cohorts. Eur. J. Epidemiol. 2018, 33, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Derkach, A.; Sampson, J.; Joseph, J.; Playdon, M.C.; Stolzenberg-Solomon, R.Z. Effects of dietary sodium on metabolites: The Dietary Approaches to Stop Hypertension (DASH)-Sodium Feeding Study. Am. J. Clin. Nutr. 2017, 106, 1131–1141. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.M.; Brinkman, P.; Weda, H.; Knobel, H.H.; Xu, Y.; Nijsen, T.M.; Goodacre, R.; Rattray, N.; Vink, T.J.; Santonico, M.; et al. Methodological considerations for large-scale breath analysis studies: Lessons from the U-BIOPRED severe asthma project. J. Breath Res. 2018, 13, 016001. [Google Scholar] [CrossRef] [Green Version]
- Kirwan, J.A.; Brennan, L.; Broadhurst, D.; Fiehn, O.; Cascante, M.; Dunn, W.B.; Schmidt, M.A.; Velagapudi, V. Preanalytical processing and biobanking procedures of biological samples for metabolomics research: A White paper, community perspective (for “Precision Medicine and Pharmacometabolomics Task Group”-the Metabolomics Society Initiative). Clin. Chem. 2018, 64, 1158–1182. [Google Scholar] [CrossRef] [Green Version]
- Breier, M.; Wahl, S.; Prehn, C.; Fugmann, M.; Ferrari, U.; Weise, M.; Banning, F.; Seissler, J.; Grallert, H.; Adamski, J.; et al. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS ONE 2014, 9, e89728. [Google Scholar] [CrossRef]
- Vaught, J.; Bledsoe, M.; Watson, P. Biobanking on multiple continents: Will international coordination follow? Biopreserv. Biobank. 2014, 12, 1–2. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. IARC Biobank. Available online: http://ibb.iarc.fr/links/index.php (accessed on 30 November 2019).
- Norwegian Institute of Public Health. Promoting Harmonization of Epidemiological Biobanks in Europe. Available online: https://www.fhi.no/en/projects/fp6-phoebe-promoting-harmonisat/ (accessed on 30 November 2019).
- Consortium, BBMRI-ERIC. Biobanking and Biomolecular Resources Research Infrastructure. Available online: http://www.bbmri-eric.eu/services/standardisation/ (accessed on 30 November 2019).
- La Frano, M.R.; Carmichael, S.L.; Ma, C.; Hardley, M.; Shen, T.; Wong, R.; Rosales, L.; Borkowski, K.; Pedersen, T.L.; Shaw, G.M.; et al. Impact of post-collection freezing delay on the reliability of serum metabolomics in samples reflecting the California mid-term pregnancy biobank. Metabolomics 2018, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Haid, M.; Muschet, C.; Wahl, S.; Römisch-Margl, W.; Prehn, C.; Möller, G.; Adamski, J. Long-term stability of human plasma metabolites during storage at −80 °C. J. Proteome Res. 2018, 17, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Betsou, F.; Cadamuro, J.; Cornes, M.; Fleischhacker, M.; Fruekilde, P.; Neumaier, M.; Nybo, M.; Padoan, A.; Plebani, M.; et al. Preanalytical challenges—Time for solutions. Clin. Chem. Lab. Med. 2019, 57, 974–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [Green Version]
- Hyotylainen, T.; Ahonen, L.; Poho, P.; Oresic, M. Lipidomics in biomedical research-practical considerations. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 800–803. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Kamleh, M.A.; Ebbels, T.M.; Spagou, K.; Masson, P.; Want, E.J. Optimizing the use of quality control samples for signal drift correction in large-scale urine metabolic profiling studies. Anal. Chem. 2012, 84, 2670–2677. [Google Scholar] [CrossRef]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 4. [Google Scholar] [CrossRef]
- Liu, X.; Hoene, M.; Wang, X.; Yin, P.; Haring, H.U.; Xu, G.; Lehmann, R. Serum or plasma, what is the difference? Investigations to facilitate the sample material selection decision making process for metabolomics studies and beyond. Anal. Chim. Acta 2018, 1037, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Kohler, I.; Hankemeier, T.; van der Graaf, P.H.; Knibbe, C.A.J.; van Hasselt, J.G.C. Integrating clinical metabolomics-based biomarker discovery and clinical pharmacology to enable precision medicine. Eur. J. Pharm. Sci. 2017, 109, S15–S21. [Google Scholar] [CrossRef] [PubMed]
- Sieber-Ruckstuhl, N.S.; Burla, B.; Spoerel, S.; Schmid, F.; Venzin, C.; Cazenave-Gassiot, A.; Bendt, A.K.; Torta, F.; Wenk, M.R.; Boretti, F.S. Changes in the canine plasma lipidome after short- and long-term excess glucocorticoid exposure. Sci. Rep. 2019, 9, 6015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beger, R.D. Interest is high in improving quality control for clinical metabolomics: Setting the path forward for community harmonization of quality control standards. Metabolomics 2018, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Plebani, M. The detection and prevention of errors in laboratory medicine. Ann. Clin. Biochem. 2009, 47, 101–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.A.; Ulmer, C.Z.; Jones, C.M.; Koelmel, J.P.; Yost, R.A. NIST lipidomics workflow questionnaire: An assessment of community-wide methodologies and perspectives. Metabolomics 2018, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khadka, M.; Todor, A.; Maner-Smith, K.M.; Colucci, J.K.; Tran, V.; Gaul, D.A.; Anderson, E.J.; Natrajan, M.S.; Rouphael, N.; Mulligan, M.J.; et al. The effect of anticoagulants, temperature, and time on the human plasma metabolome and lipidome from healthy donors as determined by liquid chromatography-mass spectrometry. Biomolecules 2019, 9, 200. [Google Scholar] [CrossRef] [Green Version]
- Robin, T.; Barnes, A.; Dulaurent, S.; Loftus, N.; Baumgarten, S.; Moreau, S.; Marquet, P.; El Balkhi, S.; Saint-Marcoux, F. Fully automated sample preparation procedure to measure drugs of abuse in plasma by liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2018, 410, 5071–5083. [Google Scholar] [CrossRef]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics. A review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Sa, M.R.; Ji, J.; Vaniya, A.; Wancewicz, B.; Roberts, B.S.; Torbašinović, H.; Lee, T.; Mehta, S.S.; et al. Structure annotation of all mass spectra in untargeted metabolomics. Anal. Chem. 2019, 91, 2155–2162. [Google Scholar] [CrossRef]
- Anton, G.; Wilson, R.; Yu, Z.H.; Prehn, C.; Zukunft, S.; Adamski, J.; Heier, M.; Meisinger, C.; Romisch-Margl, W.; Wang-Sattler, R.; et al. Pre-analytical sample quality: Metabolite ratios as an intrinsic marker for prolonged room temperature exposure of serum samples. PLoS ONE 2015, 10, e0121495. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Peter, A.; Franken, H.; Zhao, X.; Neukamm, S.S.; Rosenbaum, L.; Lucio, M.; Zell, A.; Haring, H.U.; Xu, G.; et al. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clin. Chem. 2013, 59, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Mooney, S.J.; Pejaver, V. Big data in public health: Terminology, machine learning, and privacy. Annu. Rev. Public Health 2018, 39, 95–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Livera, A.M.; Sysi-Aho, M.; Jacob, L.; Gagnon-Bartsch, J.A.; Castillo, S.; Simpson, J.A.; Speed, T.P. Statistical methods for handling unwanted variation in metabolomics data. Anal. Chem. 2015, 87, 3606–3615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soneson, C.; Gerster, S.; Delorenzi, M. Batch effect confounding leads to strong bias in performance estimates obtained by cross-validation. PLoS ONE 2014, 9, e100335. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.X.; Huang, H.C.; Begg, C.B. Cautionary note on using cross-validation for molecular classification. J. Clin. Oncol. 2016, 34, 3931–3938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Albert, F.; Llorach, R.; Garcia-Aloy, M.; Ziyatdinov, A.; Andres-Lacueva, C.; Perera, A. Intensity drift removal in LC/MS metabolomics by common variance compensation. Bioinformatics 2014, 30, 2899–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehrens, R.; Hageman, J.A.; van Eeuwijk, F.; Kooke, R.; Flood, P.J.; Wijnker, E.; Keurentjes, J.J.; Lommen, A.; van Eekelen, H.D.; Hall, R.D.; et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics 2016, 12, 88. [Google Scholar] [CrossRef] [Green Version]
- Luan, H.; Ji, F.; Chen, Y.; Cai, Z. statTarget: A streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data. Anal. Chim. Acta 2018, 1036, 66–72. [Google Scholar] [CrossRef]
- Willforss, J.; Chawade, A.; Levander, F. NormalyzerDE: Online tool for improved normalization of omics expression data and high-sensitivity differential expression analysis. J. Proteome Res. 2019, 18, 732–740. [Google Scholar] [CrossRef]
- Li, B.; Tang, J.; Yang, Q.; Li, S.; Cui, X.; Li, Y.; Chen, Y.; Xue, W.; Li, X.; Zhu, F. NOREVA: Normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017, 45, W162–W170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, W.W.B.; Wang, W.; Wong, L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 2017, 35, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.W.; Deutsch, E.W.; Au, C.E.; Kearney, R.E.; Beavis, R.; Sechi, S.; Nilsson, T.; Bergeron, J.J. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods 2009, 6, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2013, 2, 13. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A technology platform for identifying knowns and unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [Green Version]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Jarmusch, A.K.; Vargas, F.; Aksenov, A.A.; Gauglitz, J.M.; Weldon, K.; Petras, D.; da Silva, R.; Quinn, R.; Melnik, A.V.; et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Wang, L.; Xing, X.; Chen, L.; Yang, L.; Su, X.; Rabitz, H.; Lu, W.; Rabinowitz, J.D. Peak annotation and verification engine for untargeted LC-MS metabolomics. Anal. Chem. 2019, 91, 1838–1846. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Ivanisevic, J.; Thomas, A.; Sidibé, J.; Teav, T.; Guijas, C.; Aisporna, A.E.; Rinehart, D.; Hoang, L.; et al. XCMS-MRM and METLIN-MRM: A cloud library and public resource for targeted analysis of small molecules. Nat. Methods 2018, 15, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Su, M.; Qiu, Y.; Jia, W.; Du, X. ADAP-GC 3.0: Improved peak detection and deconvolution of co-eluting metabolites from GC/TOF-MS data for metabolomics studies. Anal. Chem. 2016, 88, 8802–8811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korf, A.; Jeck, V.; Schmid, R.; Helmer, P.O.; Hayen, H. Lipid species annotation at double bond position level with custom databases by extension of the mzmine 2 open-source software package. Anal. Chem. 2019, 91, 5098–5105. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MZmine 2 data-preprocessing to enhance molecular networking reliability. Anal. Chem. 2017, 89, 7836–7840. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2017, 15, 53. [Google Scholar] [CrossRef]
- Beaulieu-Jones, B.K.; Greene, C.S. Reproducibility of computational workflows is automated using continuous analysis. Nat. Biotechnol. 2017, 35, 342–346. [Google Scholar] [CrossRef]
- Chong, J.; Yamamoto, M.; Xia, J. MetaboAnalystR 2.0: From raw spectra to biological insights. Metabolites 2019, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Caron, C.; Duperier, C.; Jacob, D.; Thévenot, E.A.; Giacomoni, F.; Le Corguillé, G.; Martin, J.-F.; Tremblay-Franco, M.; Landi, M.; Pétéra, M.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2014, 31, 1493–1495. [Google Scholar] [CrossRef] [Green Version]
- Ruttkies, C.; Schober, D.; Peters, K.; Neumann, S.; Gonzalez-Beltran, A.; Izzo, M.; Rocca-Serra, P.; Sansone, S.-A.; Johnson, D.; Reed, M.A.C.; et al. PhenoMeNal: Processing and analysis of metabolomics data in the cloud. GigaScience 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Weber, R.J.M.; Lawson, T.N.; Salek, R.M.; Ebbels, T.M.D.; Glen, R.C.; Goodacre, R.; Griffin, J.L.; Haug, K.; Koulman, A.; Moreno, P.; et al. Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through Galaxy. Metabolomics 2016, 13, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armitage, E.G.; Godzien, J.; Alonso-Herranz, V.; Lopez-Gonzalvez, A.; Barbas, C. Missing value imputation strategies for metabolomics data. Electrophoresis 2015, 36, 3050–3060. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; Ruhaak, L.R.; Kelly, K.; Weiss, R.H.; Kim, K. Effects of imputation on correlation: Implications for analysis of mass spectrometry data from multiple biological matrices. Brief. Bioinform. 2017, 18, 312–320. [Google Scholar] [CrossRef] [Green Version]
- Hrydziuszko, O.; Viant, M.R. Missing values in mass spectrometry based metabolomics: An undervalued step in the data processing pipeline. Metabolomics 2012, 8, 161–174. [Google Scholar] [CrossRef]
- Jin, Z.; Yu, T.; Kang, J. Missing value imputation for LC-MS metabolomics data by incorporating metabolic network and adduct ion relations. Bioinformatics 2017, 34, 1555–1561. [Google Scholar] [CrossRef]
- Voillet, V.; Besse, P.; Liaubet, L.; San Cristobal, M.; Gonzalez, I. Handling missing rows in multi-omics data integration: Multiple imputation in multiple factor analysis framework. BMC Bioinform. 2016, 17, 402. [Google Scholar] [CrossRef] [Green Version]
- Gerl, M.J.; Klose, C.; Surma, M.A.; Fernandez, C.; Melander, O.; Mannisto, S.; Borodulin, K.; Havulinna, A.S.; Salomaa, V.; Ikonen, E.; et al. Machine learning of human plasma lipidomes for obesity estimation in a large population cohort. PLoS Biol. 2019, 17, e3000443. [Google Scholar] [CrossRef] [Green Version]
- Vvedenskaya, O.; Wang, Y.; Ackerman, J.M.; Knittelfelder, O.; Shevchenko, A. Analytical challenges in human plasma lipidomics: A winding path towards the truth. TrAC Trends Anal. Chem. 2019, 120, 115277. [Google Scholar] [CrossRef]
- Burla, B.; Arita, M.; Arita, M.; Bendt, A.K.; Cazenave-Gassiot, A.; Dennis, E.A.; Ekroos, K.; Han, X.; Ikeda, K.; Liebisch, G.; et al. MS-based lipidomics of human blood plasma: A community-initiated position paper to develop accepted guidelines. J. Lipid Res. 2018, 59, 2001–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.A.; Heckert, A.; Ulmer, C.Z.; Jones, C.M.; Koelmel, J.P.; Abdullah, L.; Ahonen, L.; Alnouti, Y.; Armando, A.M.; Asara, J.M.; et al. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J. Lipid Res. 2017, 58, 2275–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebisch, G.; Ahrends, R.; Arita, M.; Arita, M.; Bowden, J.A.; Ejsing, C.S.; Griffiths, W.J.; Holčapek, M.; Köfeler, H.; Mitchell, T.W.; et al. Lipidomics needs more standardization. Nat. Metab. 2019, 1, 745–747. [Google Scholar] [CrossRef] [Green Version]
- Rustam, Y.H.; Reid, G.E. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem. 2018, 90, 374–397. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Shi, X.; Wang, S.; Xu, G. Multidimensional liquid chromatography-mass spectrometry for metabolomic and lipidomic analyses. TrAC Trends Anal. Chem. 2019, 120, 115302. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fan, S.; Wancewicz, B.; Cajka, T.; Sa, M.; Showalter, M.R.; Baillie, R.; Tenenbaum, J.D.; Louie, G.; Kaddurah-Daouk, R.; et al. Generation and quality control of lipidomics data for the alzheimer’s disease neuroimaging initiative cohort. Sci. Data 2018, 5, 180263. [Google Scholar] [CrossRef]
- Fan, S.; Kind, T.; Cajka, T.; Hazen, S.L.; Tang, W.H.W.; Kaddurah-Daouk, R.; Irvin, M.R.; Arnett, D.K.; Barupal, D.K.; Fiehn, O. Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal. Chem. 2019, 91, 3590–3596. [Google Scholar] [CrossRef]
- Quell, J.D.; Römisch-Margl, W.; Haid, M.; Krumsiek, J.; Skurk, T.; Halama, A.; Stephan, N.; Adamski, J.; Hauner, H.; Mook-Kanamori, D.; et al. Characterization of bulk phosphatidylcholine compositions in human plasma using side-chain resolving lipidomics. Metabolites 2019, 9, 109. [Google Scholar] [CrossRef] [Green Version]
- Gathungu, R.M.; Larrea, P.; Sniatynski, M.J.; Marur, V.R.; Bowden, J.A.; Koelmel, J.P.; Starke-Reed, P.; Hubbard, V.S.; Kristal, B.S. Optimization of electrospray ionization source parameters for lipidomics to reduce misannotation of in-source fragments as precursor ions. Anal. Chem. 2018, 90, 13523–13532. [Google Scholar] [CrossRef]
- Leaptrot, K.L.; May, J.C.; Dodds, J.N.; McLean, J.A. Ion mobility conformational lipid atlas for high confidence lipidomics. Nat. Commun. 2019, 10, 985. [Google Scholar] [CrossRef]
- Lipidomic Standards Initiative. The Lipidomics Standard Initiative. Available online: https://lipidomics-standards-initiative.org/ (accessed on 30 November 2019).
- Liebisch, G.; Vizcaino, J.A.; Kofeler, H.; Trotzmuller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauling, J.K.; Hermansson, M.; Hartler, J.; Christiansen, K.; Gallego, S.F.; Peng, B.; Ahrends, R.; Ejsing, C.S. Proposal for a common nomenclature for fragment ions in mass spectra of lipids. PLoS ONE 2017, 12, e0188394. [Google Scholar] [CrossRef] [Green Version]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Liu, K.H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clair, G.; Reehl, S.; Stratton, K.G.; Monroe, M.E.; Tfaily, M.M.; Ansong, C.; Kyle, J.E. Lipid Mini-On: Mining and ontology tool for enrichment analysis of lipidomic data. Bioinformatics 2019, 35, 4507–4508. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; van de Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/web: A web-based ontology enrichment tool for lipidomic data analysis. Gigascience 2019, 8. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. New Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Trainor, P.J.; DeFilippis, A.P.; Rai, S.N. Evaluation of classifier performance for multiclass phenotype discrimination in untargeted metabolomics. Metabolites 2017, 7, 30. [Google Scholar] [CrossRef]
- Long, N.P.; Jung, K.H.; Anh, N.H.; Yan, H.H.; Nghi, T.D.; Park, S.; Yoon, S.J.; Min, J.E.; Kim, H.M.; Lim, J.H.; et al. An integrative data mining and omics-based translational model for the identification and validation of oncogenic biomarkers of pancreatic cancer. Cancers 2019, 11, 155. [Google Scholar] [CrossRef] [Green Version]
- Gosiewska, A.; Biecek, P. iBreakDown: Uncertainty of Model Explanations for Non-Additive Predictive Models. Available online: https://arxiv.org/abs/1903.11420 (accessed on 30 November 2019).
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. Available online: https://arxiv.org/abs/1602.04938 (accessed on 30 November 2019).
- Long, N.P.; Park, S.; Anh, N.H.; Min, J.E.; Yoon, S.J.; Kim, H.M.; Nghi, T.D.; Lim, D.K.; Park, J.H.; Lim, J.; et al. Efficacy of integrating a novel 16-gene biomarker panel and intelligence classifiers for differential diagnosis of rheumatoid arthritis and osteoarthritis. J. Clin. Med. 2019, 8, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Beirnaert, C.; Peeters, L.; Meysman, P.; Bittremieux, W.; Foubert, K.; Custers, D.; Van der Auwera, A.; Cuykx, M.; Pieters, L.; Covaci, A.; et al. Using expert driven machine learning to enhance dynamic metabolomics data analysis. Metabolites 2019, 9, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Breast cancer metabolomics: From analytical platforms to multivariate data analysis. A review. Metabolites 2019, 9, 102. [Google Scholar] [CrossRef] [Green Version]
- Broadhurst, D.I.; Kell, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2006, 2, 171–196. [Google Scholar] [CrossRef] [Green Version]
- Long, N.P.; Yoon, S.J.; Anh, N.H.; Nghi, T.D.; Lim, D.K.; Hong, Y.J.; Hong, S.-S.; Kwon, S.W. A systematic review on metabolomics-based diagnostic biomarker discovery and validation in pancreatic cancer. Metabolomics 2018, 14, 109. [Google Scholar] [CrossRef]
- Patti, G.J.; Tautenhahn, R.; Siuzdak, G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 2012, 7, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Considine, E.C.; Thomas, G.; Boulesteix, A.L.; Khashan, A.S.; Kenny, L.C. Critical review of reporting of the data analysis step in metabolomics. Metabolomics 2017, 14, 7. [Google Scholar] [CrossRef]
- Hicks, S.C.; Peng, R.D. Elements and Principles for Characterizing Variation between Data Analyses. Available online: https://arxiv.org/abs/1903.07639v2 (accessed on 30 November 2019).
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C.; et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015, 351, h5527. [Google Scholar] [CrossRef] [Green Version]
- Moore, H.M.; Kelly, A.B.; Jewell, S.D.; McShane, L.M.; Clark, D.P.; Greenspan, R.; Hayes, D.F.; Hainaut, P.; Kim, P.; Mansfield, E.A.; et al. Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol. 2011, 119, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Lumbreras, B.; Porta, M.; Marquez, S.; Pollan, M.; Parker, L.A.; Hernandez-Aguado, I. QUADOMICS: An adaptation of the Quality Assessment of Diagnostic Accuracy Assessment (QUADAS) for the evaluation of the methodological quality of studies on the diagnostic accuracy of ‘-omics’-based technologies. Clin. Biochem. 2008, 41, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Considine, E.; Salek, R. A tool to encourage minimum reporting guideline uptake for data analysis in metabolomics. Metabolites 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gotzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for reporting observational studies. Int. J. Surg. 2014, 12, 1495–1499. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Veenstra, T.D. Metabolomics: The final frontier? Genome Med. 2012, 4, 40. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Sailani, M.R.; Contrepois, K.; Zhou, Y.; Ahadi, S.; Leopold, S.R.; Zhang, M.J.; Rao, V.; Avina, M.; Mishra, T.; et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 2019, 569, 663–671. [Google Scholar] [CrossRef]
- Bakker, O.B.; Aguirre-Gamboa, R.; Sanna, S.; Oosting, M.; Smeekens, S.P.; Jaeger, M.; Zorro, M.; Võsa, U.; Withoff, S.; Netea-Maier, R.T.; et al. Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nat. Immunol. 2018, 19, 776–786. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype–phenotype interactions. Nat. Rev. Genet. 2015, 16, 85. [Google Scholar] [CrossRef]
- Shapiro, J.A. Revisiting the central dogma in the 21st century. Ann. N. Y. Acad. Sci. 2009, 1178, 6–28. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The need for multi-omics biomarker signatures in precision medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, B.C.; Mousavi, F.; Emili, A. Single-platform ‘multi-omic’ profiling: Unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. Mol. Omics 2018, 14, 307–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, E.; Torrey, E.F.; Guest, P.C.; Bahn, S. Biomarker discovery in human cerebrospinal fluid: The need for integrative metabolome and proteome databases. Genome Med. 2012, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Darst, B.F.; Lu, Q.; Johnson, S.C.; Engelman, C.D. Integrated analysis of genomics, longitudinal metabolomics, and Alzheimer’s risk factors among 1,111 cohort participants. Genet. Epidemiol. 2019, 43, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Adamski, J. Genome-wide association studies with metabolomics. Genome Med. 2012, 4, 34. [Google Scholar] [CrossRef]
- Schüssler-Fiorenza Rose, S.M.; Contrepois, K.; Moneghetti, K.J.; Zhou, W.; Mishra, T.; Mataraso, S.; Dagan-Rosenfeld, O.; Ganz, A.B.; Dunn, J.; Hornburg, D.; et al. A longitudinal big data approach for precision health. Nat. Med. 2019, 25, 792–804. [Google Scholar] [CrossRef]
- Yousri, N.A.; Fakhro, K.A.; Robay, A.; Rodriguez-Flores, J.L.; Mohney, R.P.; Zeriri, H.; Odeh, T.; Kader, S.A.; Aldous, E.K.; Thareja, G.; et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat. Commun. 2018, 9, 333. [Google Scholar] [CrossRef] [Green Version]
- Yet, I.; Menni, C.; Shin, S.Y.; Mangino, M.; Soranzo, N.; Adamski, J.; Suhre, K.; Spector, T.D.; Kastenmuller, G.; Bell, J.T. Genetic influences on metabolite levels: A comparison across metabolomic platforms. PLoS ONE 2016, 11, e0153672. [Google Scholar] [CrossRef] [Green Version]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahim, A.; Brunk, E.; Tan, J.; O’Brien, E.J.; Kim, D.; Szubin, R.; Lerman, J.A.; Lechner, A.; Sastry, A.; Bordbar, A.; et al. Multi-omic data integration enables discovery of hidden biological regularities. Nat. Commun. 2016, 7, 13091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More is better: Recent progress in multi-omics data integration methods. Front. Genet. 2017, 8, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappoport, N.; Shamir, R. Multi-omic and multi-view clustering algorithms: Review and cancer benchmark. Nucleic Acids Res. 2018, 46, 10546–10562. [Google Scholar] [CrossRef] [PubMed]
- McGuire, A.L.; Basford, M.; Dressler, L.G.; Fullerton, S.M.; Koenig, B.A.; Li, R.; McCarty, C.A.; Ramos, E.; Smith, M.E.; Somkin, C.P.; et al. Ethical and practical challenges of sharing data from genome-wide association studies: The eMERGE Consortium experience. Genome Res. 2011, 21, 1001–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spicer, R.A.; Steinbeck, C. A lost opportunity for science: Journals promote data sharing in metabolomics but do not enforce it. Metabolomics 2018, 14, 16. [Google Scholar] [CrossRef] [Green Version]



| Country | Year | Metabolomics/Lipidomics/Multi-omics | Clinical Field | Platform | Main Conclusions | References |
|---|---|---|---|---|---|---|
| The United Kingdom | 2011 | Steroidomics | Cancer | GC-MS | This study combined population-based steroidomics research with machine learning analysis, aiming at identifying urine steroid biomarkers for differentiating adrenocortical carcinoma from benign adenoma. | Arlt et al. [27] |
| The United States | 2011 | Metabolomics | Diabetes | LC-MS | A large nested group of 2422 normoglycemic subjects in the Framingham Offspring Study was followed for 12 years to help identify a panel of three amino acids (isoleucine, phenylalanine, and tyrosine) which might serve as novel predictors for future diabetes. | Wang et al. [28] |
| The Netherlands | 2015 | Steroidomics | Cancer | GC-MS | Urinary steroid signatures were established for the discrimination of adrenal cortical carcinoma from other adrenal conditions. | Kerkhofs et al. [29] |
| The United Kingdom | 2015 | Metabolomics | Physiological state | GC-MS and LC-MS | The Husermet project has applied untargeted MS to investigate the comprehensive hydrophilic and lipophilic metabolome of serum biospecimen obtained from the phenotyping of 1200 healthy subjects. | Dunn et al. [30] |
| The United States | 2015 | Steroidomics | Cancer | LC-MS | The novel LC-MS/MS assay used in this study enabled the examination of all estrogen metabolites for epidemiological and clinical research on hormone-related diseases. | Ziegler et al. [31] |
| Multinational | 2018 | Multi-omics | Diabetes | N/A | The Environmental Determinants of Diabetes in the Young (TEDDY) study followed more than 12,000 children to explore metabolic pathways related to type 1 diabetes. | Rewers et al. [32] |
| Multinational | 2018 | Multi-omics | Obesity | NMR and LC-MS | Along with fecal metagenomes, plasma and urine metabolome revealed molecular pathways uniting the gut microbiome and the human phenome to hepatic steatosis in two cohorts of non-diabetic obese women in the FLORINASH consortium. | Hoyles et al. [33] |
| Japan | 2018 | Metabolomics | Physiological state | CE-MS (and LC-MS) | The Tsuruoka large-scale cohort study has gathered plasma metabolomics data from more than 10,000 individuals as an innovative model for preventive medicine. | Harada et al. [34] |
| Multinational | 2019 | Metabolomics | Long-term mortality risk | NMR | From the metabolic profile of 44,168 individuals, 14 biomarkers have been associated with all-cause mortality. The combination of this set of metabolite predictors and sex considerably improves mortality risk prediction compared to traditional risk factors such as age, body mass index, systolic blood pressure, or total cholesterol. | Deelen et al. [35] |
| The United Kingdom | 2019 | Metabolomics | Parkinson’s disease | TD-GC-MS | This study applied an unbiased method, taking the use of volatile sebum metabolites to diagnose Parkinson’s disease. | Trivedi et al. [36] |
| The United Kingdom | 2019 | Metabolomics | Cardiovascular disease | NMR | This study with over 7000 participants on the metabolic profile of atherosclerosis revealed that this condition was associated with perturbations of multiple interconnected pathways related to lipid, fatty acid, and amino acid metabolism, and displayed a considerably similar model between coronary and carotid atherosclerosis. | Tzoulaki et al. [37] |
| The United States | 2019 | Metabolomics | Obesity | LC-MS | This study revealed the disturbance of the metabolome in obese versus healthy individuals. Approximately a third of compounds followed changes in body mass index, suggesting the role of metabolome profiling in participant recruitment for clinical trials related to obesity. | Cirulli et al. [38] |
| The United States | 2019 | Metabolomics | Diabetes | LC-MS | A total of 69 out of 331 plasma metabolites extracted from more than 2000 samples from the Diabetes Prevention Program were linked to type 2 diabetes regardless of treatment randomization. | Chen et al. [39] |
| Sweden | 2019 | Metabolomics | Physiological state | LC-MS | Not just stopping at the cross-sectional quantification of urinary eicosanoid metabolites, this recently published study extended the scope by focusing on the long-term repeatability and stability of the method used in order to serve the large-scale analysis of multiple cohorts. | Gomez et al. [40] |
| Phases 1 | Recommendations 2 |
|---|---|
| Pre-pre-analytical |
|
| Pre-analytical |
|
| Analytical |
|
| Post-analytical |
|
| Post-post-analytical |
|
| Section/Topic | Item | Essential Topic | STROBE [151] | TRIPOD [146] | QUADOMICs [149] | Our In-House Assessment [143] | Checklist Item 1 |
|---|---|---|---|---|---|---|---|
| 1. Title and abstract | |||||||
| Title | 1a | Yes | 1 | 1 | Specify the study design with developing and/or validating purpose, the target population, and the outcome with simple and straightforward terms in the title. | ||
| Abstract | 1b | Yes | 1 | 2 | Depending on the target journal, provide in the abstract a precise and structured summary of objectives, study design, study participants, sample size, type of samples (e.g., plasma, serum, or urine), analytical platform, predictor variables, outcome, statistical method, results, and conclusions. | ||
| 2. Introduction | |||||||
| Background | 2a | Yes | 2 | 3a | Provide the scientific and clinical background (including the diagnostic or prognostic purpose) and explain the rationale for developing and/or validating the multivariable prediction model, in regard to existing models. | ||
| Objectives | 2b | Yes | 3 | 3b | Determine the objectives and hypotheses, emphasizing if it is a development and/or validation research of the model. | ||
| 3. Methods | |||||||
| Ethical approval | 3a | Yes | Clearly report ethics committee approval and participant consent. | ||||
| Study design | 3b | Yes | 4 | 4a | Item 1 | State the study design (e.g., case-control, cohort, randomized trial, or registry data) or source of data (e.g., biobank, public database). | |
| Setting | 3c | Yes | 5 | 4b | Describe the settings, locations, and relevant dates where the data were collected, including periods of follow-up, if applicable. | ||
| Participants | 3d | Yes | 6 | 5a–5c | Item 1 and 2 | Item 2–6 | (a) Cross-sectional study: Give the inclusion and exclusion criteria, and the sources and methods of recruitment of study participants. (b) Longitudinal study: Give the inclusion and exclusion criteria, and the sources and methods of recruitment of study participants. Describe follow-up methods. Give diagnostic criteria and prior treatment, if applicable. For developing purpose, the spectrum of patients should represent those who will receive the test in practice. |
| Sample size | 3e | Yes | 10 | 8 | Explain how the sample size was determined. | ||
| Sample collection | 3f | Yes | Item 3 and 4 | Item 8 | Describe the type of samples, the procedures and timing of biological sample collection with reference to clinical factors and the methods to control metabolome changes (e.g., arterial versus venous blood, circadian oscillations, pre- and post-prandial status, the time between sampling and storage). | ||
| Sample storage | 3g | Yes | Item 5 | Item 8 | Describe the methods to control chemical and enzymatic degradation and/or interconversion. | ||
| Sample preparation | 3h | Yes | Item 5 | Item 9 | Describe the methods to control analytical errors and between-batch variations. | ||
| Data acquisition | 3i | Yes | Item 10–13 | Describe the experimental conditions, the analytical validation methods, and the number of batches of analysis. | |||
| Data preprocessing and treatment | 3j | Yes | Item 15 | Report parameters for peak detection (i.e., algorithms and acceptance criteria for valid peaks), deconvolution, alignment, and correction. Describe the methods to filter data noise, impute missing values, and correct batch effects (e.g., data-driven, internal standards-based, or QC-based normalization). | |||
| Predictors | 3k | Yes | 8 | 7a–7b | Define all variables and explain their measurements used in constructing the multivariable prediction model. | ||
| Bias | 3l | Yes | 9 | Characterize any attempts to show and solve potential sources of bias. | |||
| Missing data | 3m | Yes | 12 | 9 | Describe how missing data (e.g., samples) were handled. | ||
| Metabolite identification 2 | 3n | Yes | Item 14 | Describe the methods for metabolite identification and the level of confidence of identified compounds. Report whether a match with authentic standards has been conducted. | |||
| Multi-omics data integration | 3o | Optional | Report the multi-omics data integration, if available. Description of integration strategies, such as post-analysis data integration or simultaneous integration from different omics data types, is recommended. | ||||
| Statistical analysis and modeling | 3p | Yes | 12 | 10a–10e and 12 | Item 16 | Item 16 | Describe statistical methods, the methods to detect outliers, validation methods, and performance measures (e.g., AUC, accuracy, sensitivity, and specificity). Describe any updates of the model after the validation, if done. For the validation purpose, determine any inconsistencies from the development data. |
| Outcome | 3q | Yes | 7 | 6a–6b | Explain the assessment of outcome that is predicted by the multivariable prediction model. Report any efforts to the blind assessment of the predicted outcome. | ||
| 4. Results | |||||||
| Participants | 4a | Yes | 13 | 13a–13c | Report numbers of study participants at each stage (e.g., numbers potentially eligible, screened for eligibility, validated eligible, included in the study, finishing follow-up, and analyzed) and the number of participants with missing information for predictors and outcome. Give reasons for non-participation or exclusion at each stage. | ||
| Descriptive data | 4b | Yes | 14 | 13a–13c | Designate characteristics of study participants (e.g., baseline demographic, clinical, and socioeconomic status) and information on potential confounding factors. | ||
| Data exploratory analysis | 4c | Yes | Report exploratory data analysis (e.g., using unsupervised learning approaches). | ||||
| Model development | 4d | Yes | 15–16 | 14a–14b | Specify the number of participants and outcomes in each analysis. Report the unadjusted and adjusted potential confounders that may influence associations between each predictor and outcome. | ||
| Model interpretation | 4e | Optional | 15–16 | 15a–15b | Item 14 and 15 | Present the full prediction model to enable reproducibility for individuals. Explain how to interpret the prediction model in a human-friendly approach (e.g., LIME, iBreakDown). In case uninterpretable, indeterminate data should be reported. | |
| Model performance | 4f | Yes | 15–16 | 16 | Report measures of performance of the prediction model. Describe the effects of unbalancing (e.g., cases versus controls) to the performance of the model. | ||
| Reference standards comparison | 4g | Optional | Item 6–13 | Item 7 | Compare the constructed models with currently approved approaches (e.g., CA 19.9 for diagnosing pancreatic cancer). | ||
| Model updating | 4h | Optional | 17 | 17 | Report the results of any updates of the model. An updated model using quantitative information of the tentative biomarkers are strongly recommended. | ||
| 5. Discussion | |||||||
| Key results | 5a | Yes | 18 | Summarize critical results in harmony with study objectives. | |||
| Interpretation | 5b | Yes | 20 | 19a–19b | Give an overall evaluation of results based on study objectives, results from previous similar studies, and other related evidence. For validation purpose, discuss the results in regard to the performance of development data, and any other validation data from the public databases. | ||
| Limitations | 5c | Yes | 19 | 18 | Discuss the limitations of the study, considering sources of potential confounders, biases, and statistical uncertainty. | ||
| Implications | 5d | Yes | 21 | 20 | Discuss the potential application of the model into clinical settings and suggestions for future research and practice. | ||
| 6. Other information | |||||||
| Supplementary materials | 6a | Yes | 21 | Provide available supplementary files, such as the full study protocol and generated datasets. | |||
| Funding | 6b | Yes | 22 | 22 | Give the sources of funding and the influence of each funder on the outcome and, if applicable, for the original study from which the current paper arises. | ||
| Conflicts of interest | 6c | Yes | Clearly declare potential conflicts of interest. | ||||
| Repositories for generated data | 6d | Yes | Report the public repository for generated data concerning FAIR (Findability, Accessibility, Interoperability, and Reusability) principles 3. | ||||
| Executive commands | 6e | Optional | Report any programming code used (e.g., R code). | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, N.P.; Nghi, T.D.; Kang, Y.P.; Anh, N.H.; Kim, H.M.; Park, S.K.; Kwon, S.W. Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine. Metabolites 2020, 10, 51. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo10020051
Long NP, Nghi TD, Kang YP, Anh NH, Kim HM, Park SK, Kwon SW. Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine. Metabolites. 2020; 10(2):51. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo10020051
Chicago/Turabian StyleLong, Nguyen Phuoc, Tran Diem Nghi, Yun Pyo Kang, Nguyen Hoang Anh, Hyung Min Kim, Sang Ki Park, and Sung Won Kwon. 2020. "Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine" Metabolites 10, no. 2: 51. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo10020051