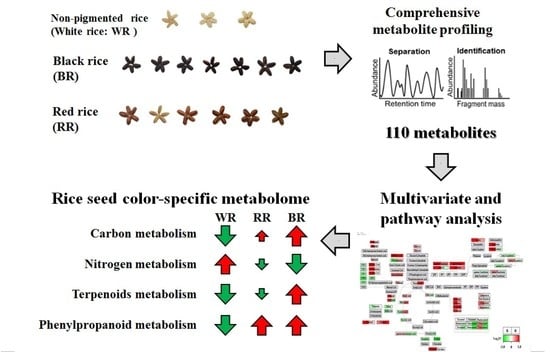
Metabolite Profiling Reveals Distinct Modulation of Complex Metabolic Networks in Non-Pigmented, Black, and Red Rice (Oryza sativa L.) Cultivars
Abstract
:1. Introduction
2. Results and Discussion
2.1. PCA, PLS-DA, and OPLS-DA
2.2. Pearson’s Correlation Analysis and HCA
2.3. PathVisio Pathway Analysis
2.4. Quality Assessment of Pigmented Rice
3. Materials and Methods
3.1. Samples and Chemicals
3.2. Extraction and Analysis of Hydrophilic Compounds
3.3. Extraction and Analysis of Lipophilic Compounds
3.3.1. Extraction and Analysis of Terpenoids and Policosanols Using GC-qMS
3.3.2. Extraction and Analysis of Fatty Acids Using GC-FID
3.3.3. Extraction and Analysis of Carotenoids Using HPLC
3.4. Extraction and Analysis of Anthocyanins
3.5. Extraction and Analysis of (+)-Catechin and (−)-Epicatechin
3.6. Headspace-SPME (HS-SPME) and Analysis of Volatile Compounds
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hussain, M.; Farooq, M.; Lee, D.J. Evaluating the role of seed priming in improving drought tolerance of pigmented and non-pigmented rice. J. Agron. Crop Sci. 2017, 203, 269–276. [Google Scholar] [CrossRef]
- Laokuldilok, T.; Shoemaker, C.F.; Jongkaewwattana, S.; Tulyathan, V. Antioxidants and antioxidant activity of several pigmented rice brans. J. Agric. Food Chem. 2011, 59, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, S.Y.; Chu, S.M.; Lim, S.H.; Suh, S.-C.; Lee, Y.-T.; Cho, H.S.; Ha, S.-H. Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J. Agric. Food Chem. 2010, 58, 12804–12809. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, V.; Ferreira, C.D.; Hoffmann, J.F.; Chaves, F.C.; Vanier, N.L.; de Oliveira, M.; Elias, M.C. Cooking quality properties and free and bound phenolics content of brown, black, and red rice grains stored at different temperatures for six months. Food Chem. 2018, 242, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Samyor, D.; Das, A.B.; Deka, S.C. Pigmented rice a potential source of bioactive compounds: A review. Int. J. Food Sci. Technol. 2017, 52, 1073–1081. [Google Scholar] [CrossRef]
- Biswas, S.K.; Kim, D.-E.; Keum, Y.-S.; Saini, R.K. Metabolite profiling and antioxidant activities of white, red, and black rice (Oryza sativa L.) grains. J. Food Meas. Charact. 2018, 12, 2484–2492. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Cros, G.; Yokota, T.; Crozier, A. Phytochemical profiles of black, red, brown, and white rice from the Camargue region of France. J. Agric. Food Chem. 2013, 61, 7976–7986. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Watanabe, S.; Crozier, A.; Fujimura, T.; Yokota, T.; Ashihara, H. Phytochemical profile of a Japanese black–purple rice. Food Chem. 2013, 141, 2821–2827. [Google Scholar] [CrossRef]
- Sukhonthara, S.; Theerakulkait, C.; Miyazawa, M. Characterization of volatile aroma compounds from red and black rice bran. J. Oleo Sci. 2009, 58, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Jun, H.I.; Shin, J.W.; Song, G.S.; Kim, Y.S. Isolation and identification of phenolic antioxidants in black rice bran. J. Food Sci. 2015, 80, C262–C268. [Google Scholar] [CrossRef]
- Kim, J.K.; Park, S.-Y.; Lim, S.-H.; Yeo, Y.; Cho, H.S.; Ha, S.-H. Comparative metabolic profiling of pigmented rice (Oryza sativa L.) cultivars reveals primary metabolites are correlated with secondary metabolites. J. Cereal Sci. 2013, 57, 14–20. [Google Scholar] [CrossRef]
- Gunaratne, A.; Wu, K.; Li, D.; Bentota, A.; Corke, H.; Cai, Y.-Z. Antioxidant activity and nutritional quality of traditional red-grained rice varieties containing proanthocyanidins. Food Chem. 2013, 138, 1153–1161. [Google Scholar] [CrossRef]
- Srisawat, U.; Panunto, W.; Kaendee, N.; Tanuchit, S.; Itharat, A.; Lerdvuthisopon, N.; Hansakul, P. Determination of phenolic compounds, flavonoids, and antioxidant activities in water extracts of Thai red and white rice cultivars. J. Med. Assoc. Thai. 2010, 93, S83–S91. [Google Scholar] [PubMed] [Green Version]
- Shen, Y.; Jin, L.; Xiao, P.; Lu, Y.; Bao, J. Total phenolics, flavonoids, antioxidant capacity in rice grain and their relations to grain color, size and weight. J. Cereal Sci. 2009, 49, 106–111. [Google Scholar] [CrossRef]
- Chumpolsri, W.; Wijit, N.; Boontakham, P.; Nimmanpipug, P.; Sookwong, P.; Luangkamin, S.; Wongpornchai, S. Variation of terpenoid flavor odorants in bran of some black and white rice varieties analyzed by GC×GC-MS. J. Food Nutr. Res. 2015, 3, 114–120. [Google Scholar] [CrossRef]
- Choi, S.; Seo, H.-S.; Lee, K.R.; Lee, S.; Lee, J. Effect of milling degrees on volatile profiles of raw and cooked black rice (Oryza sativa L. cv. Sintoheugmi). Appl. Biol. Chem. 2018, 61, 91–105. [Google Scholar] [CrossRef]
- Park, S.-Y.; Kim, J.K.; Lee, S.Y.; Oh, S.-D.; Lee, S.M.; Jang, J.-S.; Yang, C.-I.; Won, Y.-J.; Yeo, Y. Comparative analysis of phenolic acid profiles of rice grown under different regions using multivariate. Plant Omics 2014, 7, 430–437. [Google Scholar]
- Liu, Y.; Liu, J.; Liu, M.; Liu, Y.; Strappe, P.; Sun, H.; Zhou, Z. Comparative non-targeted metabolomic analysis reveals insights into the mechanism of rice yellowing. Food Chem. 2020, 308, 125621. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.; Jez, J.M. Nature’s assembly line: Biosynthesis of simple phenylpropanoids and polyketides. Plant J. 2008, 54, 750–762. [Google Scholar] [CrossRef]
- Kim, J.K.; Ha, S.-H.; Park, S.-Y.; Lee, S.M.; Kim, H.J.; Lim, S.H.; Suh, S.-C.; Kim, D.H.; Cho, H.S. Determination of lipophilic compounds in genetically modified rice using gas chromatography–time-of-flight mass spectrometry. J. Food Compos. Anal. 2012, 25, 31–38. [Google Scholar] [CrossRef]
- Choi, S.-W.; Kang, W.-W.; Osawa, T. Isolation and identification of anthocyanin pigments in black rice. Food Sci. Biotechnol. 1994, 3, 131–136. [Google Scholar]
- Yoon, H.-H.; Paik, Y.-S.; Kim, J.-B.; Hahn, T.-R. Identification of anthocyanins from Korean pigmented rice. Appl. Biol. Chem. 1995, 38, 581–583. [Google Scholar]
- Kong, L.; Wang, Y.; Cao, Y. Determination of myo-inositol and D-chiro-inositol in black rice bran by capillary electrophoresis with electrochemical detection. J. Food Compos. Anal. 2008, 21, 501–504. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Redestig, H.; Saito, K. Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 2011, 62, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.-L. Carbon and nitrogen nutrient balance signaling in plants. Plant Signal. Behav. 2009, 4, 584–591. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Hu, Z.; Yu, Y.; Mou, R.; Zhu, Z.; Beta, T. Phenolic acids, anthocyanins, proanthocyanidins, antioxidant activity, minerals and their correlations in non-pigmented, red, and black rice. Food Chem. 2018, 239, 733–741. [Google Scholar] [CrossRef]
- Hiemori, M.; Koh, E.; Mitchell, A.E. Influence of cooking on anthocyanins in black rice (Oryza sativa L. japonica var. SBR). J. Agric. Food Chem. 2009, 57, 1908–1914. [Google Scholar] [CrossRef] [PubMed]
- Chatthongpisut, R.; Schwartz, S.J.; Yongsawatdigul, J. Antioxidant activities and antiproliferative activity of Thai purple rice cooked by various methods on human colon cancer cells. Food Chem. 2015, 188, 99–105. [Google Scholar] [CrossRef]
- Kim, T.J.; Hyeon, H.; Park, N.I.; Yi, T.G.; Lim, S.-H.; Park, S.-Y.; Ha, S.-H.; Kim, J.K. A high-throughput platform for interpretation of metabolite profile data from pepper (Capsicum) fruits of 13 phenotypes associated with different fruit maturity states. Food Chem. 2020, 331, 127286. [Google Scholar] [CrossRef]
- Lister, C.E.; Lancaster, J.E.; Walker, J.R. Phenylalanine ammonia-lyase (PAL) activity and its relationship to anthocyanin and flavonoid levels in New Zealand-grown apple cultivars. J. Am. Soc. Hortic. Sci. 1996, 121, 281–285. [Google Scholar] [CrossRef]
- Wang, H.; Arakawa, O.; Motomura, Y. Influence of maturity and bagging on the relationship between anthocyanin accumulation and phenylalanine ammonia-lyase (PAL) activity in ‘Jonathan’apples. Postharvest Biol. Technol. 2000, 19, 123–128. [Google Scholar] [CrossRef]
- Bendokas, V.; Skemiene, K.; Trumbeckaite, S.; Stanys, V.; Passamonti, S.; Borutaite, V.; Liobikas, J. Anthocyanins: From plant pigments to health benefits at mitochondrial level. Crit. Rev. Food Sci. Nutr. 2020, 60, 3352–3365. [Google Scholar] [CrossRef] [PubMed]
- Martin, C. Structure, function, and regulation of the chalcone synthase. Int. Rev. Cytol. 1993, 147, 233–284. [Google Scholar]
- Hyeon, H.; Xu, J.L.; Kim, J.K.; Choi, Y. Comparative metabolic profiling of cultivated and wild black soybeans reveals distinct metabolic alterations associated with their domestication. Food Res. Int. 2020, 134, 109290. [Google Scholar] [CrossRef]
- Xu, Z.; Hua, N.; Godber, J.S. Antioxidant activity of tocopherols, tocotrienols, and γ-oryzanol components from rice bran against cholesterol oxidation accelerated by 2, 2′-azobis (2-methylpropionamidine) dihydrochloride. J. Agric. Food Chem. 2001, 49, 2077–2081. [Google Scholar] [CrossRef] [PubMed]
- Minatel, I.O.; Han, S.-I.; Aldini, G.; Colzani, M.; Matthan, N.R.; Correa, C.R.; Fecchio, D.; Yeum, K.-J. Fat-soluble bioactive components in colored rice varieties. J. Med. Food 2014, 17, 1134–1141. [Google Scholar] [CrossRef]
- Frank, T.; Reichardt, B.; Shu, Q.; Engel, K.-H. Metabolite profiling of colored rice (Oryza sativa L.) grains. J. Cereal Sci. 2012, 55, 112–119. [Google Scholar] [CrossRef]
- Kotamreddy, J.N.R.; Hansda, C.; Mitra, A. Semi-targeted metabolomic analysis provides the basis for enhanced antioxidant capacities in pigmented rice grains. J. Food Meas. Charact. 2020, 14, 1–9. [Google Scholar] [CrossRef]
- Ferri, M.; Righetti, L.; Tassoni, A. Increasing sucrose concentrations promote phenylpropanoid biosynthesis in grapevine cell cultures. J. Plant. Physiol. 2011, 168, 189–195. [Google Scholar] [CrossRef]
- Payyavula, R.S.; Navarre, D.A.; Kuhl, J.C.; Pantoja, A.; Pillai, S.S. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biol. 2012, 12, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.J.; Lee, K.B.; Baek, S.-A.; Choi, J.; Ha, S.-H.; Lim, S.-H.; Park, S.-Y.; Yeo, Y.; Park, S.U.; Kim, J.K. Determination of lipophilic metabolites for species discrimination and quality assessment of nine leafy vegetables. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 909–918. [Google Scholar] [CrossRef]
- Lee, J.H.; Ham, H.; Kim, M.Y.; Ko, J.Y.; Sim, E.-Y.; Kim, H.-J.; Lee, C.K.; Jeon, Y.H.; Jeong, H.S.; Woo, K.S. Phenolic compounds and antioxidant activity of adzuki bean cultivars. Legume Res. 2018, 41, 681–688. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.J.; Kim, S.Y.; Park, Y.J.; Lim, S.-H.; Ha, S.-H.; Park, S.U.; Lee, B.; Kim, J.K. Metabolite Profiling Reveals Distinct Modulation of Complex Metabolic Networks in Non-Pigmented, Black, and Red Rice (Oryza sativa L.) Cultivars. Metabolites 2021, 11, 367. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11060367
Kim TJ, Kim SY, Park YJ, Lim S-H, Ha S-H, Park SU, Lee B, Kim JK. Metabolite Profiling Reveals Distinct Modulation of Complex Metabolic Networks in Non-Pigmented, Black, and Red Rice (Oryza sativa L.) Cultivars. Metabolites. 2021; 11(6):367. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11060367
Chicago/Turabian StyleKim, Tae Jin, So Yeon Kim, Young Jin Park, Sun-Hyung Lim, Sun-Hwa Ha, Sang Un Park, Bumkyu Lee, and Jae Kwang Kim. 2021. "Metabolite Profiling Reveals Distinct Modulation of Complex Metabolic Networks in Non-Pigmented, Black, and Red Rice (Oryza sativa L.) Cultivars" Metabolites 11, no. 6: 367. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11060367