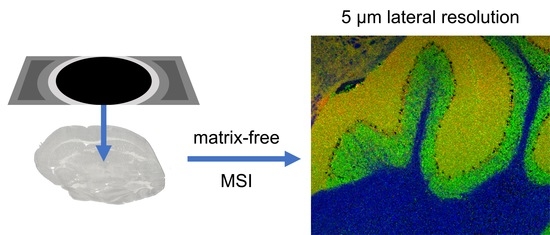
Matrix-Free High-Resolution Atmospheric-Pressure SALDI Mass Spectrometry Imaging of Biological Samples Using Nanostructured DIUTHAME Membranes
Abstract
:1. Introduction
2. Results
2.1. Desorption and Ionization Using DIUTHAME
2.2. Signal Quality and Quantity for DIUTHAME
2.3. MSI of Biological Tissues Using DIUTHAME, MALDI and LDI
2.4. DIUTHAME MSI of Tissue Sections from Various Organisms
3. Discussion
4. Materials and Methods
4.1. MSI Instrumentation
4.2. Sample Preparation
4.3. Histology
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liebl, H. Ion Microprobe Mass Analyzer. J. Appl. Phys. 1967, 38, 5277–5283. [Google Scholar] [CrossRef]
- Gilmore, I.S.; Heiles, S.; Pieterse, C.L. Metabolic Imaging at the Single-Cell Scale: Recent Advances in Mass Spectrometry Imaging. Annu. Rev. Anal. Chem. 2019, 12, 201–224. [Google Scholar] [CrossRef] [PubMed]
- Spengler, B. Mass spectrometry imaging of biomolecular information. Anal. Chem. 2015, 87, 64–82. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Biopolymers. Anal. Chem. 1991, 63, 1193A–1203A. [Google Scholar] [CrossRef]
- Spengler, B.; Hubert, M.; Kaufmann, R. MALDI ion imaging and biological ion imaging with a new scanning UV-laser microprobe. In Proceedings of the 42nd ASMS Conference on Mass Spectrometry and Allied Topics, Chicago, IL, USA, 29 May–3 June 1994; p. 1041. Available online: https://www.uni-giessen.de/fbz/fb08/Inst/iaac/spengler/forschung/dateien/poster_maldi_anwendung (accessed on 1 September 2021).
- Römpp, A.; Spengler, B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 2013, 139, 759–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturtevant, D.; Lee, Y.-J.; Chapman, K.D. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr. Opin. Biotechnol. 2016, 37, 53–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kompauer, M.; Heiles, S.; Spengler, B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 2017, 14, 90–96. [Google Scholar] [CrossRef]
- Goodwin, R.J.A. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. J. Proteomics 2012, 75, 4893–4911. [Google Scholar] [CrossRef]
- Calvano, C.D.; Monopoli, A.; Cataldi, T.R.I.; Palmisano, F. MALDI matrices for low molecular weight compounds: An endless story? Anal. Bioanal. Chem. 2018, 410, 4015–4038. [Google Scholar] [CrossRef]
- Beavis, R.C.; Chait, B.T. Cinnamic acid derivatives as matrices for ultraviolet laser desorption mass spectrometry of proteins. Rapid Commun. Mass Spectrom. 1989, 3, 432–435. [Google Scholar] [CrossRef]
- Thomas, A.; Charbonneau, J.L.; Fournaise, E.; Chaurand, P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: Enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 2012, 84, 2048–2054. [Google Scholar] [CrossRef]
- Soltwisch, J.; Jaskolla, T.W.; Hillenkamp, F.; Karas, M.; Dreisewerd, K. Ion yields in UV-MALDI mass spectrometry as a function of excitation laser wavelength and optical and physico-chemical properties of classical and halogen-substituted MALDI matrixes. Anal. Chem. 2012, 84, 6567–6576. [Google Scholar] [CrossRef] [PubMed]
- Takáts, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef] [Green Version]
- Wiseman, J.M.; Ifa, D.R.; Song, Q.; Cooks, R.G. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. Engl. 2006, 45, 7188–7192. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, R.J.A.; Takats, Z.; Bunch, J. A Critical and Concise Review of Mass Spectrometry Applied to Imaging in Drug Discovery. SLAS DISCOV. Adv. Sci. Drug Discov. 2020, 25, 963–976. [Google Scholar] [CrossRef]
- Sunner, J.; Dratz, E.; Chen, Y.C. Graphite surface-assisted laser desorption/ionization time-of-flight mass spectrometry of peptides and proteins from liquid solutions. Anal. Chem. 1995, 67, 4335–4342. [Google Scholar] [CrossRef]
- Picca, R.A.; Calvano, C.D.; Cioffi, N.; Palmisano, F. Mechanisms of Nanophase-Induced Desorption in LDI-MS. A Short Review. Nanomaterials 2017, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-C.; Shiea, J.; Sunner, J. Thin-layer chromatography–mass spectrometry using activated carbon, surface-assisted laser desorption/ionization. J. Chromatogr. A 1998, 826, 77–86. [Google Scholar] [CrossRef]
- Lo, C.-Y.; Lin, J.-Y.; Chen, W.-Y.; Chen, C.-T.; Chen, Y.-C. Surface-assisted laser desorption/ionization mass spectrometry on titania nanotube arrays. J. Am. Soc. Mass Spectrom. 2008, 19, 1014–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, E.P.; Apon, J.V.; Luo, G.; Saghatelian, A.; Daniels, R.H.; Sahi, V.; Dubrow, R.; Cravatt, B.F.; Vertes, A.; Siuzdak, G. Desorption/ionization on silicon nanowires. Anal. Chem. 2005, 77, 1641–1646. [Google Scholar] [CrossRef]
- Chen, Y.; Vertes, A. Adjustable fragmentation in laser desorption/ionization from laser-induced silicon microcolumn arrays. Anal. Chem. 2006, 78, 5835–5844. [Google Scholar] [CrossRef] [PubMed]
- McLean, J.A.; Stumpo, K.A.; Russell, D.H. Size-selected (2–10 nm) gold nanoparticles for matrix assisted laser desorption ionization of peptides. J. Am. Chem. Soc. 2005, 127, 5304–5305. [Google Scholar] [CrossRef]
- Kawasaki, H.; Yonezawa, T.; Watanabe, T.; Arakawa, R. Platinum Nanoflowers for Surface-Assisted Laser Desorption/Ionization Mass Spectrometry of Biomolecules. J. Phys. Chem. C 2007, 111, 16278–16283. [Google Scholar] [CrossRef]
- Lee, K.-H.; Chiang, C.-K.; Lin, Z.-H.; Chang, H.-T. Determining enediol compounds in tea using surface-assisted laser desorption/ionization mass spectrometry with titanium dioxide nanoparticle matrices. Rapid Commun. Mass Spectrom. 2007, 21, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kawasaki, H.; Yonezawa, T.; Arakawa, R. Surface-assisted laser desorption/ionization mass spectrometry (SALDI-MS) of low molecular weight organic compounds and synthetic polymers using zinc oxide (ZnO) nanoparticles. J. Mass Spectrom. 2008, 43, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, W.; Zhang, H.; Zhong, M.; Cao, W.; Li, Z.; Huang, X.; Nie, Z.; Liu, J.; Li, P.; et al. Polydopamine-Modified Substrates for High-Sensitivity Laser Desorption Ionization Mass Spectrometry Imaging. ACS Appl. Mater. Interfaces 2019, 11, 46140–46148. [Google Scholar] [CrossRef]
- Naito, Y.; Kotani, M.; Ohmura, T. A novel laser desorption/ionization method using through hole porous alumina membranes. Rapid Commun. Mass Spectrom. 2018, 32, 1851–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Nakamura, S.; Fouquet, T.N.J.; Ohmura, T.; Kotani, M.; Naito, Y. Simple Pretreatment for the Analysis of Additives and Polymers by Surface-Assisted Laser Desorption/Ionization Mass Spectrometry Using a Through-Hole Alumina Membrane as a Functional Substrate. J. Am. Soc. Mass Spectrom. 2020, 31, 298–307. [Google Scholar] [CrossRef]
- Kuwata, K.; Itou, K.; Kotani, M.; Ohmura, T.; Naito, Y. DIUTHAME enables matrix-free mass spectrometry imaging of frozen tissue sections. Rapid Commun. Mass Spectrom. 2020, 34, e8729. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Kotani, M.; Ohmura, T. Novel Blotting Method for Mass Spectrometry Imaging of Metabolites in Strawberry Fruit by Desorption/Ionization Using Through Hole Alumina Membrane. Foods 2020, 9, 408. [Google Scholar] [CrossRef] [Green Version]
- Law, K.P.; Larkin, J.R. Recent advances in SALDI-MS techniques and their chemical and bioanalytical applications. Anal. Bioanal. Chem. 2011, 399, 2597–2622. [Google Scholar] [CrossRef]
- Kompauer, M.; Heiles, S.; Spengler, B. AP-MALDI MSI of Lipids in Mouse Brain Tissue Sections. 2017. Available online: https://protocolexchange.researchsquare.com/article/nprot-5227/v1 (accessed on 1 September 2021).
- Müller, M.A.; Kompauer, M.; Strupat, K.; Heiles, S.; Spengler, B. Implementation of a High-Repetition-Rate Laser in an AP-SMALDI MSI System for Enhanced Measurement Performance. J. Am. Soc. Mass Spectrom. 2021, 32, 465–472. [Google Scholar] [CrossRef]
- Small, D.M. The Effects of Glyceride Structure on Absorption and Metabolism. Annu. Rev. Nutr. 1991, 11, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.R.; Wang, Q.; Friedt, W.; Spengler, B.; Gottwald, S.; Römpp, A. High resolution mass spectrometry imaging of plant tissues: Towards a plant metabolite atlas. Analyst 2015, 140, 7696–7709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, W.H.; Verdin, A.; de Pauw, E.; Malherbe, C.; Eppe, G. Surface-assisted laser desorption/ionization mass spectrometry imaging: A review. Mass Spectrom. Rev. 2020, 1–48. [Google Scholar] [CrossRef]
- Song, K.; Cheng, Q. Desorption and ionization mechanisms and signal enhancement in surface assisted laser desorption ionization mass spectrometry (SALDI-MS). Appl. Spectrosc. Rev. 2020, 55, 220–242. [Google Scholar] [CrossRef]
- Lai, S.K.-M.; Tang, H.-W.; Lau, K.-C.; Ng, K.-M. Nanosecond UV Laser Ablation of Gold Nanoparticles: Enhancement of Ion Desorption by Thermal-Driven Desorption, Vaporization, or Phase Explosion. J. Phys. Chem. C 2016, 120, 20368–20377. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.; Zhan, L.; Xue, J.; Wang, J.; Xiong, C.; Nie, Z. Hot electron transfer promotes ion production in plasmonic metal nanostructure assisted laser desorption ionization mass spectrometry. Chem. Commun. 2018, 54, 10905–10908. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-H.; Ng, K.-M. The Hidden Heroes: Holes in Charge-Driven Desorption Mass Spectrometry. Anal. Chem. 2020, 92, 5645–5649. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Miyajima, K.; Mafuné, F. Thermionic Emission of Electrons from Gold Nanoparticles by Nanosecond Pulse-Laser Excitation of Interband. J. Phys. Chem. C 2007, 111, 11246–11251. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, Z.; Wang, Y.; Teng, F.; Du, J.; Dou, S.; Lu, N. Investigation of Surface Morphology on Ion Desorption in SALDI-MS on Tailored Silicon Nanopillar Arrays. J. Phys. Chem. C 2020, 124, 2450–2457. [Google Scholar] [CrossRef]
- Paschke, C.; Leisner, A.; Hester, A.; Maass, K.; Guenther, S.; Bouschen, W.; Spengler, B. Mirion—A software package for automatic processing of mass spectrometric images. J. Am. Soc. Mass Spectrom. 2013, 24, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2006, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, A.; Phapale, P.; Chernyavsky, I.; Lavigne, R.; Fay, D.; Tarasov, A.; Kovalev, V.; Fuchser, J.; Nikolenko, S.; Pineau, C.; et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods 2017, 14, 57–60. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, M.A.; Bhandari, D.R.; Spengler, B. Matrix-Free High-Resolution Atmospheric-Pressure SALDI Mass Spectrometry Imaging of Biological Samples Using Nanostructured DIUTHAME Membranes. Metabolites 2021, 11, 624. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11090624
Müller MA, Bhandari DR, Spengler B. Matrix-Free High-Resolution Atmospheric-Pressure SALDI Mass Spectrometry Imaging of Biological Samples Using Nanostructured DIUTHAME Membranes. Metabolites. 2021; 11(9):624. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11090624
Chicago/Turabian StyleMüller, Max A., Dhaka R. Bhandari, and Bernhard Spengler. 2021. "Matrix-Free High-Resolution Atmospheric-Pressure SALDI Mass Spectrometry Imaging of Biological Samples Using Nanostructured DIUTHAME Membranes" Metabolites 11, no. 9: 624. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11090624