Maternal Nutrient Restriction Disrupts Gene Expression and Metabolites Associated with Urea Cycle, Steroid Synthesis, Glucose Homeostasis, and Glucuronidation in Fetal Calf Liver
Abstract
:1. Introduction
2. Results
2.1. Fetal Carcass Traits
2.2. Metabolomic Profile of Fetal Liver
2.3. Effect of MUN on the Gene Expression Profile of the Fetal Liver
2.4. GO Analysis of MUN-Associated Metabolic Pathways
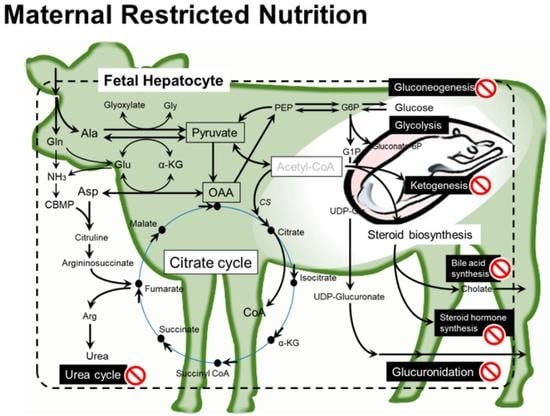
3. Discussion
3.1. Levels of Nutrients for Dams to Be Compared in the Present Study
3.2. Major Impacts of MUN on Fetal Liver
3.3. Fetal Liver Metabolites and Metabolisms Affected by MUN
3.3.1. Amino Acid Metabolism and Urea Cycle
3.3.2. Metabolisms Associated with Energy Production
3.3.3. Steroid Biosynthesis
3.3.4. Glucuronidation for Catabolism of Steroid and Drugs
3.3.5. Lipid and Fatty Acid Metabolisms
3.3.6. Other Metabolites and Gene Expression
4. Materials and Methods
4.1. Animals and Dietary Treatments
4.2. Sample Collection
4.3. Sample Preparation for CE-TOFMS
4.4. CE-TOFMS and the Data Analysis
4.5. RNA Preparation and Complementary DNA (cDNA) Synthesis
4.6. Microarray Analysis
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.8. Functional Annotation of Target Genes
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Govoni, K.E.; Reed, S.A.; Zinn, S.A. CELL BIOLOGY SYMPOSIUM: METABOLIC RESPONSES TO STRESS: FROM ANIMAL TO CELL: Poor maternal nutrition during gestation: Effects on offspring whole-body and tissue-specific metabolism in livestock species1,2. J. Anim. Sci. 2019, 97, 3142–3152. [Google Scholar] [CrossRef]
- Desai, M.; Crowther, N.J.; Lucas, A.; Hales, C.N. Organ-selective growth in the offspring of protein-restricted mothers. Br. J. Nutr. 1996, 76, 591–603. [Google Scholar] [CrossRef] [Green Version]
- Osgerby, J.C.; Wathes, D.C.; Howard, D.; Gadd, T.S. The effect of maternal undernutrition on ovine fetal growth. J. Endocrinol. 2002, 173, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Godfrey, K.M.; Costello, P.M.; Lillycrop, K.A. The developmental environment, epigenetic biomarkers and long-term health. J. Dev. Orig. Health Dis. 2015, 6, 399–406. [Google Scholar] [CrossRef]
- Lee, H.S. Impact of Maternal Diet on the Epigenome during In Utero Life and the Developmental Programming of Diseases in Childhood and Adulthood. Nutrients 2015, 7, 9492–9507. [Google Scholar] [CrossRef] [Green Version]
- Sandoval, C.; Wu, G.; Smith, S.B.; Dunlap, K.A.; Satterfield, M.C. Maternal Nutrient Restriction and Skeletal Muscle Development: Consequences for Postnatal Health. Adv. Exp. Med. Biol. 2020, 1265, 153–165. [Google Scholar] [CrossRef]
- Morrison, J.L. Sheep Models of Intrauterine Growth Restriction: Fetal Adaptations and Consequences. Clin. Exp. Pharmacol. Physiol. 2008, 35, 730–743. [Google Scholar] [CrossRef]
- Gruppuso, P.A.; Boylan, J.M.; Anand, P.; Bienieki, T.C. Effects of maternal starvation on hepatocyte proliferation in the late gestation fetal rat. Pediatr. Res. 2005, 57, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Kind, K.L.; Roberts, C.T.; Sohlstrom, A.I.; Katsman, A.; Clifton, P.M.; Robinson, J.S.; Owens, J.A. Chronic maternal feed restriction impairs growth but increases adiposity of the fetal guinea pig. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R119–R126. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, M.A.; Gopalakrishnan, G.S.; Bispham, J.; Gentili, S.; McMillen, I.C.; Rhind, S.M.; Rae, M.T.; Kyle, C.E.; Brooks, A.N.; Jones, C.; et al. Maternal nutrient restriction in early pregnancy programs hepatic mRNA expression of growth-related genes and liver size in adult male sheep. J. Endocrinol. 2007, 192, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Liu, Y.; Li, L.; Li, M.; Zhang, C.; Ao, C.; Hou, X. Effects of maternal undernutrition during late pregnancy on the development and function of ovine fetal liver. Anim. Reprod. Sci. 2014, 147, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Shibato, J.; Rakwal, R.; Saito, T.; Tamura, G.; Kuwagata, M.; Shioda, S. Seeking genes responsible for developmental origins of health and disease from the fetal mouse liver following maternal food restriction. Congenit. Anom. 2014, 54, 195–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, L.A.; Zhang, L.; Tuersunjiang, N.; Ma, Y.; Long, N.M.; Uthlaut, A.B.; Smith, D.T.; Nathanielsz, P.W.; Ford, S.P. Early maternal undernutrition programs increased feed intake, altered glucose metabolism and insulin secretion, and liver function in aged female offspring. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R795–R804. [Google Scholar] [CrossRef] [Green Version]
- Almond, K.; Bikker, P.; Lomax, M.; Symonds, M.E.; Mostyn, A. The influence of maternal protein nutrition on offspring development and metabolism: The role of glucocorticoids. Proc. Nutr. Soc. 2012, 71, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Crouse, M.S.; Caton, J.S.; Cushman, R.A.; McLean, K.J.; Dahlen, C.R.; Borowicz, P.P.; Reynolds, L.P.; Ward, A.K. Moderate nutrient restriction of beef heifers alters expression of genes associated with tissue metabolism, accretion, and function in fetal liver, muscle, and cerebrum by day 50 of gestation. Transl. Anim. Sci. 2019, 3, 855–866. [Google Scholar] [CrossRef]
- Copping, K.J.; Hernandez-Medrano, J.; Hoare, A.; Hummitzsch, K.; McMillen, I.C.; Morrison, J.L.; Rodgers, R.J.; Perry, V.E.A. Maternal periconceptional and first trimester protein restriction in beef heifers: Effects on placental parameters and fetal and neonatal calf development. Reprod. Fertil. Dev. 2020, 32, 495–507. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Guo, C.; Hu, F.; Zhu, W.; Mao, S. Undernutrition-induced lipid metabolism disorder triggers oxidative stress in maternal and fetal livers using a model of pregnant sheep. FASEB J. 2020, 34, 6508–6520. [Google Scholar] [CrossRef] [Green Version]
- Lillycrop, K.A.; Rodford, J.; Garratt, E.S.; Slater-Jefferies, J.L.; Godfrey, K.M.; Gluckman, P.D.; Hanson, M.A.; Burdge, G.C. Maternal protein restriction with or without folic acid supplementation during pregnancy alters the hepatic transcriptome in adult male rats. Br. J. Nutr. 2010, 103, 1711–1719. [Google Scholar] [CrossRef] [Green Version]
- Lecoutre, S.; Montel, V.; Vallez, E.; Pourpe, C.; Delmont, A.; Eury, E.; Verbanck, M.; Dickes-Coopman, A.; Daubersies, P.; Lesage, J.; et al. Transcription profiling in the liver of undernourished male rat offspring reveals altered lipid metabolism pathways and predisposition to hepatic steatosis. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E1094–E1107. [Google Scholar] [CrossRef]
- Xue, Y.; Guo, C.; Hu, F.; Zhu, W.; Mao, S. Maternal undernutrition induces fetal hepatic lipid metabolism disorder and affects the development of fetal liver in a sheep model. FASEB J. 2019, 33, 9990–10004. [Google Scholar] [CrossRef]
- Muroya, S.; Zhang, Y.; Kinoshita, A.; Otomaru, K.; Oshima, K.; Gotoh, Y.; Oshima, I.; Sano, M.; Roh, S.; Oe, M.; et al. Maternal Undernutrition during Pregnancy Alters Amino Acid Metabolism and Gene Expression Associated with Energy Metabolism and Angiogenesis in Fetal Calf Muscle. Metabolites 2021, 11, 582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Otomaru, K.; Oshima, K.; Goto, Y.; Oshima, I.; Muroya, S.; Sano, M.; Saneshima, R.; Nagao, Y.; Kinoshita, A.; et al. Effects of low and high levels of maternal nutrition consumed for the entirety of gestation on the development of muscle, adipose tissue, bone, and the organs of Wagyu cattle fetuses. Anim. Sci. J. 2021, 92, e13600. [Google Scholar] [CrossRef]
- Li, C.; Schlabritz-Loutsevitch, N.E.; Hubbard, G.B.; Han, V.; Nygard, K.; Cox, L.A.; McDonald, T.J.; Nathanielsz, P.W. Effects of maternal global nutrient restriction on fetal baboon hepatic insulin-like growth factor system genes and gene products. Endocrinology 2009, 150, 4634–4642. [Google Scholar] [CrossRef] [PubMed]
- Paradis, F.; Wood, K.M.; Swanson, K.C.; Miller, S.P.; McBride, B.W.; Fitzsimmons, C. Maternal nutrient restriction in mid-to-late gestation influences fetal mRNA expression in muscle tissues in beef cattle. BMC Genom. 2017, 18, 632. [Google Scholar] [CrossRef] [PubMed]
- National Agriculture and Food Research Organization. Japanese Feeding Standard for Beef Cattle 2008 edn; Japan Livestock Industry Association: Tokyo, Japan, 2009. (In Japanese) [Google Scholar]
- Takimoto, E.; Fukushima, N.; Kanetani, T.; Nishimura, S. The Optimum Method for Additional Daily Nutrient Requirements for Pregnant Japanese Black Cattle in Later Pregnancy. Bull. Okayama Prefect. Cent. Anim. Husb. Res. 2019, 9, 11–16. (In Japanese) [Google Scholar]
- Reynolds, C.K. Metabolism of Nitrogenous Compounds by Ruminant Liver. J. Nutr. 1992, 122, 850–854. [Google Scholar] [CrossRef]
- Chandel, N.S. Navigating Metabolism; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Mew, N.A.; Pappa, M.B.; Gropman, A.L. Chapter 57-Urea Cycle Disorders. In Rosenberg’s Molecular and Genetic Basis of Neurological and Psychiatric Disease, 5th ed.; Rosenberg, R.N., Pascual, J.M., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 633–647. [Google Scholar] [CrossRef]
- Chandel, N.S. Amino Acid Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040584. [Google Scholar] [CrossRef]
- Seal, C.J.; Reynolds, C.K. Nutritional Implications of Gastrointestinal and Liver Metabolism in Ruminants. Nutr. Res. Rev. 1993, 6, 185–208. [Google Scholar] [CrossRef] [Green Version]
- Danfær, A. Nutrient metabolism and utilization in the liver. Livest. Prod. Sci. 1994, 39, 115–127. [Google Scholar] [CrossRef]
- Chandel, N.S. Carbohydrate Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040568. [Google Scholar] [CrossRef]
- Miyazaki, M.; Kato, M.; Tanaka, K.; Tanaka, M.; Kohjima, M.; Nakamura, K.; Enjoji, M.; Nakamuta, M.; Kotoh, K.; Takayanagi, R. Increased hepatic expression of dipeptidyl peptidase-4 in non-alcoholic fatty liver disease and its association with insulin resistance and glucose metabolism. Mol. Med. Rep. 2012, 5, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Aroor, A.R.; Habibi, J.; Ford, D.A.; Nistala, R.; Lastra, G.; Manrique, C.; Dunham, M.M.; Ford, K.D.; Thyfault, J.P.; Parks, E.J.; et al. Dipeptidyl peptidase-4 inhibition ameliorates Western diet-induced hepatic steatosis and insulin resistance through hepatic lipid remodeling and modulation of hepatic mitochondrial function. Diabetes 2015, 64, 1988–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumeier, C.; Schlüter, L.; Saussenthaler, S.; Laeger, T.; Rödiger, M.; Alaze, S.A.; Fritsche, L.; Häring, H.U.; Stefan, N.; Fritsche, A.; et al. Elevated hepatic DPP4 activity promotes insulin resistance and non-alcoholic fatty liver disease. Mol. Metab. 2017, 6, 1254–1263. [Google Scholar] [CrossRef]
- Schulz, K.; Frahm, J.; Kersten, S.; Meyer, U.; Rehage, J.; Piechotta, M.; Meyerholz, M.; Breves, G.; Reiche, D.; Sauerwein, H.; et al. Effects of Inhibiting Dipeptidyl Peptidase-4 (DPP4) in Cows with Subclinical Ketosis. PLoS ONE 2015, 10, e0136078. [Google Scholar] [CrossRef] [PubMed]
- Chartrel, N.; Picot, M.; El Medhi, M.; Arabo, A.; Berrahmoune, H.; Alexandre, D.; Maucotel, J.; Anouar, Y.; Prévost, G. The Neuropeptide 26RFa (QRFP) and Its Role in the Regulation of Energy Homeostasis: A Mini-Review. Front. Neurosci. 2016, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Allerton, T.D.; Primeaux, S.D. QRFP-26 enhances insulin’s effects on glucose uptake in rat skeletal muscle cells. Peptides 2015, 69, 77–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulumba, M.; Jossart, C.; Granata, R.; Gallo, D.; Escher, E.; Ghigo, E.; Servant, M.J.; Marleau, S.; Ong, H. GPR103b functions in the peripheral regulation of adipogenesis. Mol. Endocrinol. 2010, 24, 1615–1625. [Google Scholar] [CrossRef] [Green Version]
- Lie, S.; Morrison, J.L.; Williams-Wyss, O.; Suter, C.M.; Humphreys, D.T.; Ozanne, S.E.; Zhang, S.; MacLaughlin, S.M.; Kleemann, D.O.; Walker, S.K.; et al. Impact of embryo number and maternal undernutrition around the time of conception on insulin signaling and gluconeogenic factors and microRNAs in the liver of fetal sheep. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1013–E1024. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Yang, H.; Yan, Q.; Ren, A.; Kong, Z.; Tang, S.; Han, X.; Tan, Z.; Salem, A.Z.M. Evidence for liver energy metabolism programming in offspring subjected to intrauterine undernutrition during midgestation. Nutr. Metab. 2019, 16, 20. [Google Scholar] [CrossRef] [Green Version]
- Chandel, N.S. Lipid Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040576. [Google Scholar] [CrossRef]
- O’Shaughnessy, P.J.; Monteiro, A.; Bhattacharya, S.; Fraser, M.J.; Fowler, P.A. Steroidogenic enzyme expression in the human fetal liver and potential role in the endocrinology of pregnancy. Mol. Hum. Reprod. 2013, 19, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Diczfalusy, E. Steroid metabolism in the human foeto-placental unit. Acta Endocrinol. 1969, 61, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 1998, 101, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.R. Extra-adrenal regeneration of glucocorticoids by 11beta-hydroxysteroid dehydrogenase type 1: Physiological regulator and pharmacological target for energy partitioning. Proc. Nutr. Soc. 2007, 66, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, S.A.; McCabe, E.L.; Gathercole, L.L.; Hassan-Smith, Z.K.; Larner, D.P.; Bujalska, I.J.; Stewart, P.M.; Tomlinson, J.W.; Lavery, G.G. 11β-HSD1 is the major regulator of the tissue-specific effects of circulating glucocorticoid excess. Proc. Natl. Acad. Sci. USA 2014, 111, E2482–E2491. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhou, X.; Yang, H.; Gebeyew, K.; Yan, Q.; Zhou, C.; He, Z.; Tan, Z. Transcriptome analysis reveals liver metabolism programming in kids from nutritional restricted goats during mid-gestation. PeerJ 2021, 9, e10593. [Google Scholar] [CrossRef]
- Tukey, R.H.; Strassburg, C.P. Human UDP-glucuronosyltransferases: Metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 581–616. [Google Scholar] [CrossRef]
- Burchell, B.; Brierley, C.H.; Monaghan, G.; Clarke, D.J. The Structure and Function of the UDP-Glucuronosyltransferase Gene Family. In Advances in Pharmacology; Goldstein, D.S., Eisenhofer, G., McCarty, R., Eds.; Academic Press: Cambridge, MA, USA, 1997; Volume 42, pp. 335–338. [Google Scholar]
- Ono, M.; Ohtaki, T.; Nakahashi, T.; Tsumagari, S. Effect of feed restriction on hepatic estradiol metabolism and liver function in cows. J. Vet. Med. Sci. 2019, 81, 1873–1878. [Google Scholar] [CrossRef]
- Osabe, M.; Sugatani, J.; Fukuyama, T.; Ikushiro, S.; Ikari, A.; Miwa, M. Expression of hepatic UDP-glucuronosyltransferase 1A1 and 1A6 correlated with increased expression of the nuclear constitutive androstane receptor and peroxisome proliferator-activated receptor alpha in male rats fed a high-fat and high-sucrose diet. Drug Metab. Dispos. 2008, 36, 294–302. [Google Scholar] [CrossRef]
- Allen, M.S.; Bradford, B.J.; Oba, M. Board Invited Review: The hepatic oxidation theory of the control of feed intake and its application to ruminants. J. Anim. Sci. 2009, 87, 3317–3334. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Suárez, Y. ANGPTL4: A multifunctional protein involved in metabolism and vascular homeostasis. Curr. Opin. Hematol. 2020, 27, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Kong, Y.; Peng, D.-q. New insights into apolipoprotein A5 in controlling lipoprotein metabolism in obesity and the metabolic syndrome patients. Lipids Health Dis. 2018, 17, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muroya, S.; Ogasawara, H.; Nohara, K.; Oe, M.; Ojima, K.; Hojito, M. Coordinated alteration of mRNA-microRNA transcriptomes associated with exosomes and fatty acid metabolism in adipose tissue and skeletal muscle in grazing cattle. Asian-Australas J. Anim. Sci. 2020, 33, 1824–1836. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, L.; Berbée, J.F.; van Dijk, S.J.; van Dijk, K.W.; Bensadoun, A.; Kema, I.P.; Voshol, P.J.; Müller, M.; Rensen, P.C.; Kersten, S. Angptl4 Upregulates Cholesterol Synthesis in Liver via Inhibition of LPL- and HL-Dependent Hepatic Cholesterol Uptake. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2420–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teratani, T.; Tomita, K.; Wada, A.; Sugihara, N.; Higashiyama, M.; Inaba, K.; Horiuchi, K.; Hanawa, Y.; Nishii, S.; Mizoguchi, A.; et al. Angiopoietin-like protein 4 deficiency augments liver fibrosis in liver diseases such as nonalcoholic steatohepatitis in mice through enhanced free cholesterol accumulation in hepatic stellate cells. Hepatol. Res. 2021, 51, 580–592. [Google Scholar] [CrossRef]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential Metabolomics Reveals Ophthalmic Acid as an Oxidative Stress Biomarker Indicating Hepatic Glutathione Consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migliaccio, V.; Lionetti, L.; Putti, R.; Scudiero, R. Exposure to Dichlorodiphenyldichloroethylene (DDE) and Metallothionein Levels in Rats Fed with Normocaloric or High-Fat Diet: A Review. Int. J. Mol. Sci. 2020, 21, 1903. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, H.; Kakimoto, Y.; Nakajima, T.; Kanazawa, A.; Sano, I. Isolation and identification of 2-aminoethylphosphonic acid from bovine brain. Nature 1965, 207, 1197–1198. [Google Scholar] [CrossRef]
- Alhadeff, J.A.; Van Bruggen, J.T.; Doyle Daves, G. Biosynthetic studies on 2-aminoethylphosphonic acid in a mammalian (rat) system. Biochim. Biophys. Acta (BBA) Gen. Subj. 1972, 286, 103–106. [Google Scholar] [CrossRef]
- Hasegawa, S.; Tamari, M.; Kametaka, M. Isolation of diacylglyceryl-2-aminoethylphosphonate from bovine liver. J. Biochem. 1976, 80, 531–535. [Google Scholar] [CrossRef]
- Joseph, J.C.; Henderson, T.O. 2-Aminoethylphosphonic acid metabolism in the rat. Lipids 1977, 12, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Kemp, P.; Dawson, R.M.C.; Klein, R.A. A new bacterial sphingophospholipid containing 3-aminopropane-1,2-diol. Biochem. J. 1972, 130, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Institute of Laboratory Animal Science Research Support Center Kagoshima University. The Guide for the Care and Use of Experimental Animals. Available online: https://www.kufm.kagoshima-u.ac.jp/~animal/tebiki.htm (accessed on 27 January 2022). (In Japanese).





| LN (n = 6) | HN (n = 5) | ||||
|---|---|---|---|---|---|
| Mean * | SE * | Mean | SE | p-Value | |
| Age (d) | 260.7 | 1.6 | 261.6 | 1.5 | 0.684 |
| BW (kg) | 23.4 | 2.2 | 32.5 | 0.5 | 0.005 |
| Liver (g) | 487.3 | 41.8 | 627.6 | 19.8 | 0.020 |
| %BW | 2.1 | 0.07 | 1.93 | 0.03 | 0.071 |
| Compound | LN (n = 4) | HN (n = 4) | Ratio (LN/HN) | p-Value | ||
|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | |||
| Aspartate | 16.500 | 0.559 | 11.750 | 0.545 | 1.40 | 0.002 |
| 4-Amino-3-hydroxybutyrate | 0.009 | 0.001 | 0.017 | 0.001 | 0.55 | 0.012 |
| 2-Aminoethylphosphonate | 0.043 | 0.005 | 0.097 | 0.013 | 0.44 | 0.013 |
| Betaine aldehyde | 0.088 | 0.008 | 0.045 | 0.007 | 1.95 | 0.014 |
| UDP-glucose/UDP-galactose | 0.133 | 0.014 | 0.225 | 0.019 | 0.59 | 0.015 |
| N5-Ethylglutamine | 0.320 | 0.022 | 0.558 | 0.057 | 0.57 | 0.015 |
| UDP-glucuronate | 0.248 | 0.017 | 0.335 | 0.016 | 0.74 | 0.019 |
| 2-Hydroxybutyrate | 0.061 | 0.021 | 0.163 | 0.022 | 0.38 | 0.028 |
| Glycerol | 7.925 | 0.828 | 4.925 | 0.399 | 1.61 | 0.030 |
| 3-Aminopropane-1,2-diol | 0.017 | 0.001 | 0.007 | 0.003 | 2.62 | 0.032 |
| Alanine | 31.750 | 1.431 | 26.500 | 0.829 | 1.20 | 0.033 |
| Octanoate | 0.002 | 0.000 | 0.013 | 0.003 | 0.19 | 0.035 |
| 6-Phosphogluconate | 0.173 | 0.017 | 0.102 | 0.017 | 1.70 | 0.044 |
| Gly-Leu | 0.021 | 0.001 | 0.029 | 0.002 | 0.72 | 0.046 |
| Ophthalmate | 14.250 | 1.244 | 6.875 | 2.280 | 2.07 | 0.049 |
| Pantothenate | 0.170 | 0.009 | 0.230 | 0.022 | 0.74 | 0.071 |
| N6,N6,N6-Trimethyllysine | 0.130 | 0.009 | 0.103 | 0.006 | 1.26 | 0.084 |
| γ-Glu-Ser | 0.072 | 0.006 | 0.049 | 0.008 | 1.48 | 0.085 |
| Methionine sulfoxide | 0.007 | 0.003 | 0.019 | 0.005 | 0.36 | 0.091 |
| Gluconate | 0.308 | 0.020 | 0.400 | 0.035 | 0.77 | 0.092 |
| Metabolism/Pathway | Hits/Total Metabolites | p-Value | Increased in LN | Decreased in LN |
|---|---|---|---|---|
| Urea Cycle | 2/29 | 0.0013 | Asp, Ala | |
| Starch and Sucrose Metabolism | 3/31 | 0.0014 | Glucuronate, UDP-Glc, UDP-glucuronate | |
| Malate-Aspartate Shuttle | 1/10 | 0.0019 | Asp | |
| Nucleotide Sugars Metabolism | 3/20 | 0.0024 | UDP-Gal, UDP-Glc, UDP-glucuronate | |
| Arginine and Proline Metabolism | 3/53 | 0.0034 | Asp | FAD, Phosphocreatine |
| Propanoate Metabolism | 3/42 | 0.0035 | CoA | FAD, 2-HBA |
| Glutamate Metabolism | 5/49 | 0.0036 | Asp, Ala, CoA | FAD, GSSG |
| Beta-Alanine Metabolism | 5/34 | 0.0050 | Asp, His, CoA | FAD, Pantothenate |
| Tyrosine Metabolism | 2/72 | 0.0065 | Asp | FAD |
| Purine Metabolism | 2/74 | 0.0065 | Asp | FAD |
| Aspartate Metabolism | 2/35 | 0.0065 | Asp | FAD |
| Porphyrin Metabolism | 2/40 | 0.0070 | FAD, UDP-glucuronate | |
| Galactose Metabolism | 3/38 | 0.0072 | Glycerol | UDP-Gal, UDP-Glc |
| Betaine Metabolism | 4/21 | 0.0076 | BTL | Dimethylglycine, FAD, Met |
| Glycerolipid Metabolism | 3/25 | 0.0095 | CoA, Glycerol | FAD |
| Warburg Effect | 3/58 | 0.0116 | 6-Phosphogluconate, CoA | FAD |
| Pantothenate and CoA Biosynthesis | 3/21 | 0.0138 | CMP, CoA | Pantothenate |
| Lactose Synthesis | 2/20 | 0.0148 | UDP-Gal, UDP-Glc | |
| Sphingolipid Metabolism | 2/40 | 0.0170 | Ser | UDP-Glc |
| Androgen and Estrogen Metabolism | 1/33 | 0.0186 | UDP-glucuronate |
| Category | Term | p-Value | Fold Enrichment | Differentially Expressed Genes ** |
|---|---|---|---|---|
| KEGG Pathway | ||||
| bta01100:Metabolic pathways | <0.001 | 1.698 | ENO3, G6PC, PIPOX, HSD11B1, ADH4, ASL, PCK1, FDPS, CPS1, TAT, HMGCS2, UGT2A1, ARG2, UGT1A1, ACOX2, EHHADH, ALDOC, FBP1, SAO, HSD17B6, UGT1A6, ASS1 | |
| bta01130:Biosynthesis of antibiotics | <0.001 | 2.647 | TAT, ENO3, ASL, HMGCS2, PCK1, FDPS, ARG2, ASS1, EHHADH, ALDOC, FBP1 | |
| bta00010:Glycolysis/Gluconeogenesis | <0.001 | 3.463 | G6PC, ENO3, ADH4, ALDOC, PCK1, FBP1 | |
| bta05204:Chemical carcinogenesis | <0.001 | 3.289 | UGT1A1, HSD11B1, ADH4, UGT2A1, UGT1A6 | |
| bta03320:PPAR signaling pathway | <0.001 | 3.289 | APOA5, FADS2, ACOX2, EHHADH, ANGPTL4, PCK1 | |
| bta04146:Peroxisome | <0.001 | 2.885 | PIPOX, ACOX2, EHHADH | |
| bta04974:Protein digestion and absorption | <0.001 | 2.885 | PRCP, DPP4, COL1A1, COL1A2, | |
| bta00350:Tyrosine metabolism | <0.001 | 3.939 | SAO, ADH4, TAT | |
| bta00980:Metabolism of xenobiotics by cytochrome P450 | <0.001 | 3.286 | UGT1A1, HSD11B1, ADH4, UGT2A1, UGT1A6 | |
| bta00071:Fatty acid degradation | <0.001 | 3.843 | ADH4, EHHADH | |
| GO: Biological Process | ||||
| GO:0006695~cholesterol biosynthetic process | <0.001 | 7.442 | FDPS, APOA5, HMGCS2 | |
| GO:0006958~complement activation, classical pathway | <0.001 | 5.953 | ||
| GO:0055089~fatty acid homeostasis | <0.001 | 7.938 | ||
| GO:0002548~monocyte chemotaxis | <0.001 | 4.106 | CCL14, CCL21, CCL5 | |
| GO:0071346~cellular response to interferon-gamma | <0.001 | 3.638 | CCL14, CCL21, CCL5 | |
| GO:0055114~oxidation-reduction process | <0.001 | 1.696 | SAO, HSD17B6, FADS2, PIPOX, | |
| GO:0042593~glucose homeostasis | <0.001 | 2.492 | G6PC, PRCP | |
| GO:0070098~chemokine-mediated signaling pathway | <0.001 | 3.247 | CCL14, CCL21, CCL5 | |
| GO:0000050~urea cycle | 0.001 | 8.505 | ARG2, CPS1, ASL, ASS1 | |
| GO:0030593~neutrophil chemotaxis | 0.001 | 3.040 | CCL14, CCL21, CCL5 | |
| Category | Term | p-Value | Fold Enrichment | Differentially Expressed Genes ** |
|---|---|---|---|---|
| KEGG Pathway | ||||
| bta03008:Ribosome biogenesis in eukaryotes | <0.001 | 4.8155 | ||
| bta03013:RNA transport | <0.001 | 2.3926 | ||
| bta00970:Aminoacyl-tRNA biosynthesis | <0.001 | 3.9018 | ||
| bta05230:Central carbon metabolism in cancer | <0.001 | 3.0680 | FGFR2 | |
| bta04141:Protein processing in endoplasmic reticulum | 0.001 | 2.0779 | ||
| bta04066:HIF-1 signaling pathway | 0.005 | 2.2862 | ||
| bta05200:Pathways in cancer | 0.013 | 1.4705 | FGFR2 | |
| bta05031:Amphetamine addiction | 0.015 | 2.4022 | PPP1R1B, ATF4 | |
| bta04010:MAPK signaling pathway | 0.025 | 1.5492 | FGFR2, ATF4 | |
| bta04978:Mineral absorption | 0.028 | 2.6603 | MT2A, MT1A, MT1E | |
| GO: Biological Process | ||||
| GO:0006364~rRNA processing | <0.001 | 4.6384 | RRP9 | |
| GO:0006457~protein folding | <0.001 | 2.9962 | ||
| GO:0000462~maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | <0.001 | 5.0379 | ||
| GO:0042742~defense response to bacterium | <0.001 | 2.8497 | CATHL4, CATHL3 | |
| GO:0008542~visual learning | 0.001 | 3.6639 | PPP1R1B | |
| GO:0051085~chaperone mediated protein folding requiring cofactor | 0.002 | 8.0148 | ||
| GO:0042254~ribosome biogenesis | 0.002 | 4.1036 | ||
| GO:0008380~RNA splicing | 0.004 | 2.5647 | ||
| GO:0045926~negative regulation of growth | 0.005 | 6.4119 | MT2A, MT1A | |
| GO:0000470~maturation of LSU-rRNA | 0.006 | 4.8089 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muroya, S.; Zhang, Y.; Otomaru, K.; Oshima, K.; Oshima, I.; Sano, M.; Roh, S.; Ojima, K.; Gotoh, T. Maternal Nutrient Restriction Disrupts Gene Expression and Metabolites Associated with Urea Cycle, Steroid Synthesis, Glucose Homeostasis, and Glucuronidation in Fetal Calf Liver. Metabolites 2022, 12, 203. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12030203
Muroya S, Zhang Y, Otomaru K, Oshima K, Oshima I, Sano M, Roh S, Ojima K, Gotoh T. Maternal Nutrient Restriction Disrupts Gene Expression and Metabolites Associated with Urea Cycle, Steroid Synthesis, Glucose Homeostasis, and Glucuronidation in Fetal Calf Liver. Metabolites. 2022; 12(3):203. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12030203
Chicago/Turabian StyleMuroya, Susumu, Yi Zhang, Kounosuke Otomaru, Kazunaga Oshima, Ichiro Oshima, Mitsue Sano, Sanggun Roh, Koichi Ojima, and Takafumi Gotoh. 2022. "Maternal Nutrient Restriction Disrupts Gene Expression and Metabolites Associated with Urea Cycle, Steroid Synthesis, Glucose Homeostasis, and Glucuronidation in Fetal Calf Liver" Metabolites 12, no. 3: 203. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12030203